Comparative proteomic analysis reveals novel insights into the interaction between rice and Xanthomonas oryzae pv. oryzae
- PMID: 33317452
- PMCID: PMC7734852
- DOI: 10.1186/s12870-020-02769-7
Comparative proteomic analysis reveals novel insights into the interaction between rice and Xanthomonas oryzae pv. oryzae
Abstract
Background: Bacterial blight, which is caused by Xanthomonas oryzae pv. oryzae (Xoo), is a devastating rice disease worldwide. Rice introgression line H471, derived from the recurrent parent Huang-Hua-Zhan (HHZ) and the donor parent PSBRC28, exhibits broad-spectrum resistance to Xoo, including to the highly virulent Xoo strain PXO99A, whereas its parents are susceptible to PXO99A. To characterize the responses to Xoo, we compared the proteome profiles of the host and pathogen in the incompatible interaction (H471 inoculated with PXO99A) and the compatible interaction (HHZ inoculated with PXO99A).
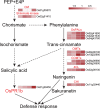
Results: In this study, a total of 374 rice differentially abundant proteins (DAPs) and 117 Xoo DAPs were detected in the comparison between H471 + PXO99A and HHZ + PXO99A. Most of the Xoo DAPs related to pathogen virulence, including the outer member proteins, type III secretion system proteins, TonB-dependent receptors, and transcription activator-like effectors, were less abundant in the incompatible interaction than in the compatible interaction. The rice DAPs were mainly involved in secondary metabolic processes, including phenylalanine metabolism and the biosynthesis of flavonoids and phenylpropanoids. Additionally, some DAPs involved in the phenolic phytoalexin and salicylic acid (SA) biosynthetic pathways accumulated much more in H471 than in HHZ after the inoculation with PXO99A, suggesting that phytoalexin and SA productions were induced faster in H471 than in HHZ. Further analyses revealed that the SA content increased much more rapidly in H471 than in HHZ after the inoculation, suggesting that the SA signaling pathway was activated faster in the incompatible interaction than in the compatible interaction.
Conclusions: Overall, our results indicate that during an incompatible interaction between H471 and PXO99A, rice plants prevent pathogen invasion and also initiate multi-component defense responses that inhibit disease development.
Keywords: Compatible interaction; Incompatible interaction; Quantitative proteome; Rice; Xanthomonas oryzae pv. oryzae.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Nino-Liu DO, Ronald PC, Bogdanove AJ. Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol Plant Pathol. 2006;7(5):303–24. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

