CircRNA_0000392 promotes colorectal cancer progression through the miR-193a-5p/PIK3R3/AKT axis
- PMID: 33317596
- PMCID: PMC7735421
- DOI: 10.1186/s13046-020-01799-1
CircRNA_0000392 promotes colorectal cancer progression through the miR-193a-5p/PIK3R3/AKT axis
Abstract
Background: Circular RNAs (circRNAs), important members of the noncoding RNA family, have been recently revealed to play a role in the pathogenic progression of diseases, particularly in the malignant progression of cancer. With the application of high-throughput sequencing technology, a large number of circRNAs have been identified in tumor tissues, and some circRNAs have been demonstrated to act as oncogenes. In this study, we analyzed the circRNA expression profile in colorectal cancer (CRC) tissues and normal adjacent tissues by high-throughput sequencing. We focused on circRNA_0000392, a circRNA with significantly increased expression in CRCtissues, and further investigated its function in the progression of colorectal cancer.
Methods: The expression profile of circRNAs in 6 pairs of CRC tissues and normal adjacent tissues was analyzed by RNA sequencing. We verified the identified differentially expressed circRNAs in additional samples by qRT-PCR and selected circRNA_0000392 to evaluate its associations with clinicopathological features. Then, we knocked down circRNA_0000392 in CRC cells and investigated the in vitro and in vivo effects using functional experiments. Dual luciferase and RNA pull-down assays were performed to further explore the downstream potential molecular mechanisms.
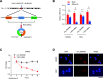
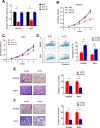
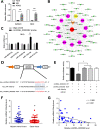
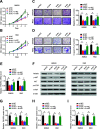
Results: CircRNA_0000392 was significantly upregulated in CRC compared with normal adjacent tissues and cell lines. The expression level of circRNA_0000392 was positively correlated with the malignant progression of CRC. Functional studies revealed that reducing the expression of circRNA_0000392 could inhibit the proliferation and invasion of CRC both in vitro and in vivo. Mechanistically, circRNA_0000392 could act as a sponge of miR-193a-5p and regulate the expression of PIK3R3, affecting the activation of the AKT-mTOR pathway in CRC cells.
Conclusions: CircRNA_0000392 functions as an oncogene through the miR-193a-5p/PIK3R3/Akt axis in CRC cells, suggesting that circRNA_0000392 is a potential therapeutic target for the treatment of colorectal cancer and a predictive marker for CRC patients.
Keywords: CircRNA_0000392; Circular RNAs (circRNAs); Colorectal cancer; PIK3R3; RNA sequencing; miR-193a-5p.
Conflict of interest statement
The authors declare that there was no conflict of interest.
Figures




Similar articles
-
The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1α translation.Mol Cancer. 2020 Nov 23;19(1):164. doi: 10.1186/s12943-020-01272-9. Mol Cancer. 2020. PMID: 33225938 Free PMC article.
-
CircRNA hsa_circ_0002577 accelerates endometrial cancer progression through activating IGF1R/PI3K/Akt pathway.J Exp Clin Cancer Res. 2020 Aug 26;39(1):169. doi: 10.1186/s13046-020-01679-8. J Exp Clin Cancer Res. 2020. PMID: 32847606 Free PMC article.
-
Circular RNA circ_0007142 regulates cell proliferation, apoptosis, migration and invasion via miR-455-5p/SGK1 axis in colorectal cancer.Anticancer Drugs. 2021 Jan 1;32(1):22-33. doi: 10.1097/CAD.0000000000000992. Anticancer Drugs. 2021. PMID: 32889894
-
Crosstalk between circRNAs and the PI3K/AKT signaling pathway in cancer progression.Signal Transduct Target Ther. 2021 Nov 24;6(1):400. doi: 10.1038/s41392-021-00788-w. Signal Transduct Target Ther. 2021. PMID: 34815385 Free PMC article. Review.
-
Comprehensive landscape and future perspectives of circular RNAs in colorectal cancer.Mol Cancer. 2021 Feb 3;20(1):26. doi: 10.1186/s12943-021-01318-6. Mol Cancer. 2021. PMID: 33536039 Free PMC article. Review.
Cited by
-
Circ_0005927 Inhibits the Progression of Colorectal Cancer by Regulating miR-942-5p/BATF2 Axis.Cancer Manag Res. 2021 Mar 11;13:2295-2306. doi: 10.2147/CMAR.S281377. eCollection 2021. Cancer Manag Res. 2021. PMID: 33732022 Free PMC article.
-
circZC3HAV1 Regulates TBC1D9 to Affect the Biological Behavior of Colorectal Cancer Cells.Biomed Res Int. 2022 Sep 16;2022:7386946. doi: 10.1155/2022/7386946. eCollection 2022. Biomed Res Int. 2022. Retraction in: Biomed Res Int. 2024 Mar 20;2024:9892656. doi: 10.1155/2024/9892656. PMID: 36164444 Free PMC article. Retracted.
-
MicroRNAs targeted mTOR as therapeutic agents to improve radiotherapy outcome.Cancer Cell Int. 2024 Jul 4;24(1):233. doi: 10.1186/s12935-024-03420-3. Cancer Cell Int. 2024. PMID: 38965615 Free PMC article. Review.
-
CircSETD2 inhibits YAP1 by interaction with HuR during breast cancer progression.Cancer Biol Ther. 2023 Dec 31;24(1):2246205. doi: 10.1080/15384047.2023.2246205. Cancer Biol Ther. 2023. PMID: 37606201 Free PMC article.
-
circASS1 overexpression inhibits the proliferation, invasion and migration of colorectal cancer cells by regulating the miR-1269a/VASH1 axis.Exp Ther Med. 2021 Oct;22(4):1155. doi: 10.3892/etm.2021.10589. Epub 2021 Aug 10. Exp Ther Med. 2021. PMID: 34504600 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

