Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID-19
- PMID: 33317616
- PMCID: PMC7734467
- DOI: 10.1186/s13054-020-03398-0
Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID-19
Abstract
Background: COVID-19 can course with respiratory and extrapulmonary disease. SARS-CoV-2 RNA is detected in respiratory samples but also in blood, stool and urine. Severe COVID-19 is characterized by a dysregulated host response to this virus. We studied whether viral RNAemia or viral RNA load in plasma is associated with severe COVID-19 and also to this dysregulated response.
Methods: A total of 250 patients with COVID-19 were recruited (50 outpatients, 100 hospitalized ward patients and 100 critically ill). Viral RNA detection and quantification in plasma was performed using droplet digital PCR, targeting the N1 and N2 regions of the SARS-CoV-2 nucleoprotein gene. The association between SARS-CoV-2 RNAemia and viral RNA load in plasma with severity was evaluated by multivariate logistic regression. Correlations between viral RNA load and biomarkers evidencing dysregulation of host response were evaluated by calculating the Spearman correlation coefficients.
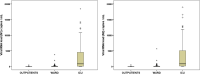
Results: The frequency of viral RNAemia was higher in the critically ill patients (78%) compared to ward patients (27%) and outpatients (2%) (p < 0.001). Critical patients had higher viral RNA loads in plasma than non-critically ill patients, with non-survivors showing the highest values. When outpatients and ward patients were compared, viral RNAemia did not show significant associations in the multivariate analysis. In contrast, when ward patients were compared with ICU patients, both viral RNAemia and viral RNA load in plasma were associated with critical illness (OR [CI 95%], p): RNAemia (3.92 [1.183-12.968], 0.025), viral RNA load (N1) (1.962 [1.244-3.096], 0.004); viral RNA load (N2) (2.229 [1.382-3.595], 0.001). Viral RNA load in plasma correlated with higher levels of chemokines (CXCL10, CCL2), biomarkers indicative of a systemic inflammatory response (IL-6, CRP, ferritin), activation of NK cells (IL-15), endothelial dysfunction (VCAM-1, angiopoietin-2, ICAM-1), coagulation activation (D-Dimer and INR), tissue damage (LDH, GPT), neutrophil response (neutrophils counts, myeloperoxidase, GM-CSF) and immunodepression (PD-L1, IL-10, lymphopenia and monocytopenia).
Conclusions: SARS-CoV-2 RNAemia and viral RNA load in plasma are associated with critical illness in COVID-19. Viral RNA load in plasma correlates with key signatures of dysregulated host responses, suggesting a major role of uncontrolled viral replication in the pathogenesis of this disease.
Keywords: COVID-19; Cytokine; ICU; Plasma; Rnaemia; SARS-CoV-2; Sepsis; Viral RNA load.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU) [Internet]. https://coronavirus.jhu.edu/map.html
-
- Chen X, Zhao B, Qu Y, Chen Y, Xiong J, Feng Y, et al. Detectable Serum Severe Acute Respiratory Syndrome Coronavirus 2 Viral Load (RNAemia) Is Closely Correlated With Drastically Elevated Interleukin 6 Level in Critically Ill Patients With Coronavirus Disease 2019. Clin Infect Dis [Internet]. [cited 2020 Jul 6]; https://academic.oup.com/cid/article/doi/10.1093/cid/ciaa449/5821311 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

