The Ubiquitin Ligase TRAIP: Double-Edged Sword at the Replisome
- PMID: 33317933
- PMCID: PMC7856240
- DOI: 10.1016/j.tcb.2020.11.007
The Ubiquitin Ligase TRAIP: Double-Edged Sword at the Replisome
Abstract
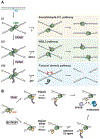
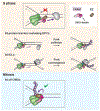
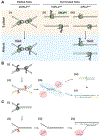
In preparation for cell division, the genome must be copied with high fidelity. However, replisomes often encounter obstacles, including bulky DNA lesions caused by reactive metabolites and chemotherapeutics, as well as stable nucleoprotein complexes. Here, we discuss recent advances in our understanding of TRAIP, a replisome-associated E3 ubiquitin ligase that is mutated in microcephalic primordial dwarfism. In interphase, TRAIP helps replisomes overcome DNA interstrand crosslinks and DNA-protein crosslinks, whereas in mitosis it triggers disassembly of all replisomes that remain on chromatin. We describe a model to explain how TRAIP performs these disparate functions and how they help maintain genome integrity.
Keywords: DNA interstrand crosslink repair; DNA-protein crosslink repair; E3 ubiquitin ligase; mitotic DNA replication; replication termination.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

