Isolation of a member of the candidate phylum 'Atribacteria' reveals a unique cell membrane structure
- PMID: 33318506
- PMCID: PMC7736352
- DOI: 10.1038/s41467-020-20149-5
Isolation of a member of the candidate phylum 'Atribacteria' reveals a unique cell membrane structure
Abstract
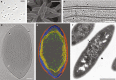
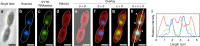
A key feature that differentiates prokaryotic cells from eukaryotes is the absence of an intracellular membrane surrounding the chromosomal DNA. Here, we isolate a member of the ubiquitous, yet-to-be-cultivated phylum 'Candidatus Atribacteria' (also known as OP9) that has an intracytoplasmic membrane apparently surrounding the nucleoid. The isolate, RT761, is a subsurface-derived anaerobic bacterium that appears to have three lipid membrane-like layers, as shown by cryo-electron tomography. Our observations are consistent with a classical gram-negative structure with an additional intracytoplasmic membrane. However, further studies are needed to provide conclusive evidence for this unique intracellular structure. The RT761 genome encodes proteins with features that might be related to the complex cellular structure, including: N-terminal extensions in proteins involved in important processes (such as cell-division protein FtsZ); one of the highest percentages of transmembrane proteins among gram-negative bacteria; and predicted Sec-secreted proteins with unique signal peptides. Physiologically, RT761 primarily produces hydrogen for electron disposal during sugar degradation, and co-cultivation with a hydrogen-scavenging methanogen improves growth. We propose RT761 as a new species, Atribacter laminatus gen. nov. sp. nov. and a new phylum, Atribacterota phy. nov.
Conflict of interest statement
The authors declare no competing interests.
Figures
Comment in
-
Cultivation of elusive microbes unearthed exciting biology.Nat Commun. 2021 Jan 4;12(1):75. doi: 10.1038/s41467-020-20393-9. Nat Commun. 2021. PMID: 33398002 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

