Distortion of the bilayer and dynamics of the BAM complex in lipid nanodiscs
- PMID: 33318620
- PMCID: PMC7736308
- DOI: 10.1038/s42003-020-01419-w
Distortion of the bilayer and dynamics of the BAM complex in lipid nanodiscs
Abstract
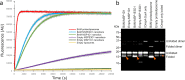
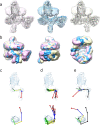
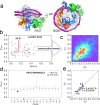
The β-barrel assembly machinery (BAM) catalyses the folding and insertion of β-barrel outer membrane proteins (OMPs) into the outer membranes of Gram-negative bacteria by mechanisms that remain unclear. Here, we present an ensemble of cryoEM structures of the E. coli BamABCDE (BAM) complex in lipid nanodiscs, determined using multi-body refinement techniques. These structures, supported by single-molecule FRET measurements, describe a range of motions in the BAM complex, mostly localised within the periplasmic region of the major subunit BamA. The β-barrel domain of BamA is in a 'lateral open' conformation in all of the determined structures, suggesting that this is the most energetically favourable species in this bilayer. Strikingly, the BAM-containing lipid nanodisc is deformed, especially around BAM's lateral gate. This distortion is also captured in molecular dynamics simulations, and provides direct structural evidence for the lipid 'disruptase' activity of BAM, suggested to be an important part of its functional mechanism.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- 108466/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- 208395/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- BB/M01151/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/N007603/1 /BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 105220/Z/14/Z/WT_/Wellcome Trust/United Kingdom
- BB/N017307/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/P018491/1/MRC_/Medical Research Council/United Kingdom
- BB/P000037/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 108466/WT_/Wellcome Trust/United Kingdom
- BB/T000635/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 208395/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

