An unbiased approach to defining bona fide cancer neoepitopes that elicit immune-mediated cancer rejection
- PMID: 33320837
- PMCID: PMC7843235
- DOI: 10.1172/JCI142823
An unbiased approach to defining bona fide cancer neoepitopes that elicit immune-mediated cancer rejection
Abstract
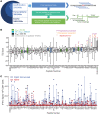
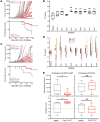
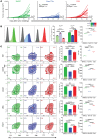
Identification of neoepitopes that are effective in cancer therapy is a major challenge in creating cancer vaccines. Here, using an entirely unbiased approach, we queried all possible neoepitopes in a mouse cancer model and asked which of those are effective in mediating tumor rejection and, independently, in eliciting a measurable CD8 response. This analysis uncovered a large trove of effective anticancer neoepitopes that have strikingly different properties from conventional epitopes and suggested an algorithm to predict them. It also revealed that our current methods of prediction discard the overwhelming majority of true anticancer neoepitopes. These results from a single mouse model were validated in another antigenically distinct mouse cancer model and are consistent with data reported in human studies. Structural modeling showed how the MHC I-presented neoepitopes had an altered conformation, higher stability, or increased exposure to T cell receptors as compared with the unmutated counterparts. T cells elicited by the active neoepitopes identified here demonstrated a stem-like early dysfunctional phenotype associated with effective responses against viruses and tumors of transgenic mice. These abundant anticancer neoepitopes, which have not been tested in human studies thus far, can be exploited for generation of personalized human cancer vaccines.
Keywords: Antigen; Bioinformatics; Cancer immunotherapy; Immunology; Oncology.
Conflict of interest statement
Figures



References
-
- Sette A, et al. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J Immunol. 1994;153(12):5586–5592. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

