Locus-specific paramutation in Zea mays is maintained by a PICKLE-like chromodomain helicase DNA-binding 3 protein controlling development and male gametophyte function
- PMID: 33320854
- PMCID: PMC7837471
- DOI: 10.1371/journal.pgen.1009243
Locus-specific paramutation in Zea mays is maintained by a PICKLE-like chromodomain helicase DNA-binding 3 protein controlling development and male gametophyte function
Abstract
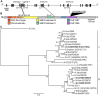
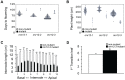
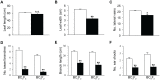
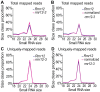
Paramutations represent directed and meiotically-heritable changes in gene regulation leading to apparent violations of Mendelian inheritance. Although the mechanism and evolutionary importance of paramutation behaviors remain largely unknown, genetic screens in maize (Zea mays) identify five components affecting 24 nucleotide RNA biogenesis as required to maintain repression of a paramutant purple plant1 (pl1) allele. Currently, the RNA polymerase IV largest subunit represents the only component also specifying proper development. Here we identify a chromodomain helicase DNA-binding 3 (CHD3) protein orthologous to Arabidopsis (Arabidopsis thaliana) PICKLE as another component maintaining both pl1 paramutation and normal somatic development but without affecting overall small RNA biogenesis. In addition, genetic tests show this protein contributes to proper male gametophyte function. The similar mutant phenotypes documented in Arabidopsis and maize implicate some evolutionarily-conserved gene regulation while developmental defects associated with the two paramutation mutants are largely distinct. Our results show that a CHD3 protein responsible for normal plant ontogeny and sperm transmission also helps maintain meiotically-heritable epigenetic regulatory variation for specific alleles. This finding implicates an intersection of RNA polymerase IV function and nucleosome positioning in the paramutation process.
Conflict of interest statement
All rmr12 materials and their uses are covered by U.S. patent 8134047 assigned to The Regents of the University of California.
Figures


Similar articles
-
Maize RNA polymerase IV defines trans-generational epigenetic variation.Plant Cell. 2013 Mar;25(3):808-19. doi: 10.1105/tpc.112.107680. Epub 2013 Mar 19. Plant Cell. 2013. PMID: 23512852 Free PMC article.
-
Trans-Homolog Interactions Facilitating Paramutation in Maize.Plant Physiol. 2015 Aug;168(4):1226-36. doi: 10.1104/pp.15.00591. Epub 2015 Jul 6. Plant Physiol. 2015. PMID: 26149572 Free PMC article. Review.
-
required to maintain repression2 is a novel protein that facilitates locus-specific paramutation in maize.Plant Cell. 2012 May;24(5):1761-75. doi: 10.1105/tpc.112.097618. Epub 2012 May 4. Plant Cell. 2012. PMID: 22562610 Free PMC article.
-
Rmr6 maintains meiotic inheritance of paramutant states in Zea mays.Genetics. 2005 Oct;171(2):725-40. doi: 10.1534/genetics.105.045260. Epub 2005 Jul 14. Genetics. 2005. PMID: 16020780 Free PMC article.
-
Paramutation: a process for acquiring trans-generational regulatory states.Curr Opin Plant Biol. 2011 Apr;14(2):210-6. doi: 10.1016/j.pbi.2011.02.005. Epub 2011 Mar 17. Curr Opin Plant Biol. 2011. PMID: 21420347 Review.
Cited by
-
RNA-directed DNA methylation prevents rapid and heritable reversal of transposon silencing under heat stress in Zea mays.PLoS Genet. 2021 Jun 14;17(6):e1009326. doi: 10.1371/journal.pgen.1009326. eCollection 2021 Jun. PLoS Genet. 2021. PMID: 34125827 Free PMC article.
-
Transcribed enhancer sequences are required for maize p1 paramutation.Genetics. 2024 Jan 3;226(1):iyad178. doi: 10.1093/genetics/iyad178. Genetics. 2024. PMID: 38169343 Free PMC article.
-
Paramutation at the maize pl1 locus is associated with RdDM activity at distal tandem repeats.PLoS Genet. 2024 May 30;20(5):e1011296. doi: 10.1371/journal.pgen.1011296. eCollection 2024 May. PLoS Genet. 2024. PMID: 38814980 Free PMC article.
-
Preliminary Evidence of a Horizontal Transfer of Paramutation Phenomenon at the pl1 Gene in Maize (Zea mays L.).Plants (Basel). 2024 Dec 24;14(1):11. doi: 10.3390/plants14010011. Plants (Basel). 2024. PMID: 39795272 Free PMC article.
-
CHROMOMETHYLTRANSFERASE3/KRYPTONITE maintains the sulfurea paramutation in Solanum lycopersicum.Proc Natl Acad Sci U S A. 2022 Mar 29;119(13):e2112240119. doi: 10.1073/pnas.2112240119. Epub 2022 Mar 24. Proc Natl Acad Sci U S A. 2022. PMID: 35324329 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

