The Role of Protein Disorder in Nuclear Transport and in Its Subversion by Viruses
- PMID: 33321790
- PMCID: PMC7764567
- DOI: 10.3390/cells9122654
The Role of Protein Disorder in Nuclear Transport and in Its Subversion by Viruses
Abstract
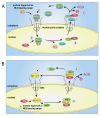
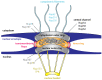
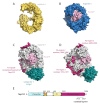
The transport of host proteins into and out of the nucleus is key to host function. However, nuclear transport is restricted by nuclear pores that perforate the nuclear envelope. Protein intrinsic disorder is an inherent feature of this selective transport barrier and is also a feature of the nuclear transport receptors that facilitate the active nuclear transport of cargo, and the nuclear transport signals on the cargo itself. Furthermore, intrinsic disorder is an inherent feature of viral proteins and viral strategies to disrupt host nucleocytoplasmic transport to benefit their replication. In this review, we highlight the role that intrinsic disorder plays in the nuclear transport of host and viral proteins. We also describe viral subversion mechanisms of the host nuclear transport machinery in which intrinsic disorder is a feature. Finally, we discuss nuclear import and export as therapeutic targets for viral infectious disease.
Keywords: nuclear export inhibitors; nuclear export sequence; nuclear import inhibitors; nuclear import sequence; nuclear transport receptors; nucleoporins; protein intrinsic disorder; viral infection.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

