Alterins Produced by Oyster-Associated Pseudoalteromonas Are Antibacterial Cyclolipopeptides with LPS-Binding Activity
- PMID: 33321943
- PMCID: PMC7764243
- DOI: 10.3390/md18120630
Alterins Produced by Oyster-Associated Pseudoalteromonas Are Antibacterial Cyclolipopeptides with LPS-Binding Activity
Abstract
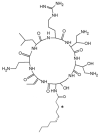
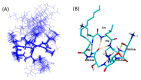
Discovery after discovery, host-associated microbiota reveal a growing list of positive effects on host homeostasis by contributing to host nutrition, improving hosts' immune systems and protecting hosts against pathogens. In that context, a collection of oyster associated bacteria producing antibacterial compounds have been established to evaluate their role in non-host-derived immunity. Here, we described alterins; potent anti-Gram negative compounds produced by Pseudoalteromonas hCg-6 and hCg-42 isolated from different healthy oyster hemolymph. The strains hCg-6 and hCg-42 produce a set of at least seven antibacterial compounds, ranging from 926 to 982 Da structurally characterized as cyclolipopeptides (CLPs). Alterins share the same cationic heptapeptidic cycle connected via an amido bond to different hydrophobic hydrocarbon tails. Their MICs disclosed a potent antibacterial activity directed against Gram-negative bacteria including oyster and human pathogens that may confer a beneficial defense mechanism to the host but also represents an untapped source of new antibiotics. The alterins' mechanisms of action have been deciphered: after binding to lipopolysaccharides (LPS), alterins provoke a membrane depolarization and permeabilization leading to bacterial lysis. As hCg-6 and hCg-42 produced a set of natural derivatives, the structure/activity relationship linked to the carbon tail is clarified. We showed that the hydrocarbon tail determines the LPS-binding properties of alterins and consequently their antibacterial activities. Its length and saturation seem to play a major role in this interaction.
Keywords: Pseudoalteromonas; alterin; antibiotic; cyclolipopeptides.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

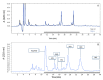
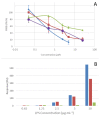
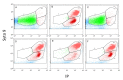
 ) and cells exhibiting an altered membrane with low (
) and cells exhibiting an altered membrane with low ( ) and high (
) and high ( ) nucleic acid content.
) nucleic acid content.References
-
- Theis K.R., Dheilly N.M., Klassen J.L., Brucker R.M., Baines J.F., Bosch T.C.G., Cryan J.F., Gilbert S.F., Goodnight C.J., Lloyd E.A., et al. Getting the Hologenome Concept Right: An Eco-Evolutionary Framework for Hosts and Their Microbiomes. mSystems. 2016;1:e00028-16. doi: 10.1128/mSystems.00028-16. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

