Perspective in Alternative Splicing Coupled to Nonsense-Mediated mRNA Decay
- PMID: 33321981
- PMCID: PMC7764535
- DOI: 10.3390/ijms21249424
Perspective in Alternative Splicing Coupled to Nonsense-Mediated mRNA Decay
Abstract
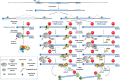
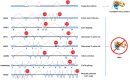
Alternative splicing (AS) of precursor mRNA (pre-mRNA) is a cellular post-transcriptional process that generates protein isoform diversity. Nonsense-mediated RNA decay (NMD) is an mRNA surveillance pathway that recognizes and selectively degrades transcripts containing premature translation-termination codons (PTCs), thereby preventing the production of truncated proteins. Nevertheless, NMD also fine-tunes the gene expression of physiological mRNAs encoding full-length proteins. Interestingly, around one third of all AS events results in PTC-containing transcripts that undergo NMD. Numerous studies have reported a coordinated action between AS and NMD, in order to regulate the expression of several genes, especially those coding for RNA-binding proteins (RBPs). This coupling of AS to NMD (AS-NMD) is considered a gene expression tool that controls the ratio of productive to unproductive mRNA isoforms, ultimately degrading PTC-containing non-functional mRNAs. In this review, we focus on the mechanisms underlying AS-NMD, and how this regulatory process is able to control the homeostatic expression of numerous RBPs, including splicing factors, through auto- and cross-regulatory feedback loops. Furthermore, we discuss the importance of AS-NMD in the regulation of biological processes, such as cell differentiation. Finally, we analyze interesting recent data on the relevance of AS-NMD to human health, covering its potential roles in cancer and other disorders.
Keywords: AS-NMD; alternative splicing (AS); gene expression regulation; nonsense-mediated RNA decay (NMD).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Burge C.S., Tuschl T., Sharp P.A. The RNA World. 2nd ed. Center for Cancer Research and Department of Biology Massachusetts Institute of Technology; Cambridge, MA, USA: 1999. Splicing of Precursors to mRNAs by the Spliceosomes.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

