NCBI's Virus Discovery Codeathon: Building "FIVE" -The Federated Index of Viral Experiments API Index
- PMID: 33322070
- PMCID: PMC7764237
- DOI: 10.3390/v12121424
NCBI's Virus Discovery Codeathon: Building "FIVE" -The Federated Index of Viral Experiments API Index
Abstract
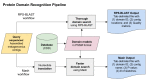
Viruses represent important test cases for data federation due to their genome size and the rapid increase in sequence data in publicly available databases. However, some consequences of previously decentralized (unfederated) data are lack of consensus or comparisons between feature annotations. Unifying or displaying alternative annotations should be a priority both for communities with robust entry representation and for nascent communities with burgeoning data sources. To this end, during this three-day continuation of the Virus Hunting Toolkit codeathon series (VHT-2), a new integrated and federated viral index was elaborated. This Federated Index of Viral Experiments (FIVE) integrates pre-existing and novel functional and taxonomy annotations and virus-host pairings. Variability in the context of viral genomic diversity is often overlooked in virus databases. As a proof-of-concept, FIVE was the first attempt to include viral genome variation for HIV, the most well-studied human pathogen, through viral genome diversity graphs. As per the publication of this manuscript, FIVE is the first implementation of a virus-specific federated index of such scope. FIVE is coded in BigQuery for optimal access of large quantities of data and is publicly accessible. Many projects of database or index federation fail to provide easier alternatives to access or query information. To this end, a Python API query system was developed to enhance the accessibility of FIVE.
Keywords: CRISPR; HIV-1; data federation; genome graphs; metagenomics; protein domain; virus.
Conflict of interest statement
J.M.C and A.R.G have received travel awards and bursaries from Oxford Nanopore Technologies, Oxford, UK. This material should not be interpreted as representing the viewpoint of the U.S. Department of Health and Human Services, the National Institutes of Health, Food and Drug Administration, National Library of Medicine, National Center for Biotechnology Information, Center for Information Technology. No other competing interests to disclose.
Figures


References
-
- SRA Database Growth. [(accessed on 3 December 2020)]; Available online: https://www.ncbi.nlm.nih.gov/sra/docs/sragrowth/
-
- Connor R., Brister R., Buchmann J., Deboutte W., Edwards R., Martí-Carreras J., Tisza M., Zalunin V., Andrade-Martínez J., Cantu A., et al. NCBI’s Virus Discovery Hackathon: Engaging Research Communities to Identify Cloud Infrastructure Requirements. Genes (Basel). 2019;10:714. doi: 10.3390/genes10090714. - DOI - PMC - PubMed
-
- STRIDES Initiative. [(accessed on 3 December 2020)]; Available online: https://datascience.nih.gov/strides.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

