Fungal Dysbiosis Correlates with the Development of Tumor-Induced Cachexia in Mice
- PMID: 33322197
- PMCID: PMC7770573
- DOI: 10.3390/jof6040364
Fungal Dysbiosis Correlates with the Development of Tumor-Induced Cachexia in Mice
Abstract
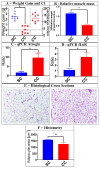
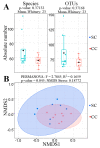
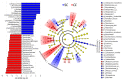
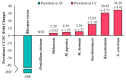
Cachexia (CC) is a devastating metabolic syndrome associated with a series of underlying diseases that greatly affects life quality and expectancy among cancer patients. Studies involving mouse models, in which CC was induced through inoculation with tumor cells, originally suggested the existence of a direct correlation between the development of this syndrome and changes in the relative proportions of several bacterial groups present in the digestive tract. However, these analyses have focus solely on the characterization of bacterial dysbiosis, ignoring the possible existence of changes in the relative populations of fungi, during the development of CC. Thus, the present study sought to expand such analyses, by characterizing changes that occur in the gut fungal population (mycobiota) of mice, during the development of cancer-induced cachexia. Our results confirm that cachectic animals, submitted to Lewis lung carcinoma (LLC) transplantation, display significant differences in their gut mycobiota, when compared to healthy controls. Moreover, identification of dysbiotic fungi showed remarkable consistency across successive levels of taxonomic hierarchy. Many of these fungi have also been associated with dysbioses observed in a series of gut inflammatory diseases, such as obesity, colorectal cancer (CRC), myalgic encephalomyelitis (ME) and inflammatory bowel disease (IBD). Nonetheless, the dysbiosis verified in the LLC model of cancer cachexia seems to be unique, presenting features observed in both obesity (reduced proportion of Mucoromycota) and CRC/ME/IBD (increased proportions of Sordariomycetes, Saccharomycetaceae and Malassezia). One species of Mucoromycota (Rhyzopus oryzae) stands out as a promising probiotic candidate in adjuvant therapies, aimed at treating and/or preventing the development of CC.
Keywords: NGS; cachexia; microbiome; microbiota; mycobiota; next generation sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Bindels L.B., Beck R., Schakman O., Martin J.C., De Backer F., Sohet F.M., Dewulf E.M., Pachikian B.D., Neyrinck A.M., Thissen J.P., et al. Restoring Specific Lactobacilli Levels Decreases Inflammation and Muscle Atrophy Markers in an Acute Leukemia Mouse Model. PLoS ONE. 2012;7:e37971. doi: 10.1371/journal.pone.0037971. - DOI - PMC - PubMed
-
- Bindels L.B., Neyrinck A.M., Claus S.P., Le Roy C.I., Grangette C., Pot B., Martinez I., Walter J., Cani P.D., Delzenne N.M. Synbiotic approach restores intestinal homeostasis and prolongs survival in leukaemic mice with cachexia. ISME J. 2016;10:1456–1470. doi: 10.1038/ismej.2015.209. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

