Molecular Insights into Determinants of Translational Readthrough and Implications for Nonsense Suppression Approaches
- PMID: 33322589
- PMCID: PMC7764779
- DOI: 10.3390/ijms21249449
Molecular Insights into Determinants of Translational Readthrough and Implications for Nonsense Suppression Approaches
Abstract
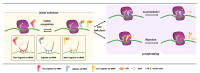
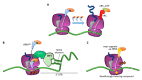
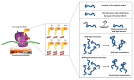
The fidelity of protein synthesis, a process shaped by several mechanisms involving specialized ribosome regions and external factors, ensures the precise reading of sense and stop codons. However, premature termination codons (PTCs) arising from mutations may, at low frequency, be misrecognized and result in PTC suppression, named ribosome readthrough, with production of full-length proteins through the insertion of a subset of amino acids. Since some drugs have been identified as readthrough inducers, this fidelity drawback has been explored as a therapeutic approach in several models of human diseases caused by nonsense mutations. Here, we focus on the mechanisms driving translation in normal and aberrant conditions, the potential fates of mRNA in the presence of a PTC, as well as on the results obtained in the research of efficient readthrough-inducing compounds. In particular, we describe the molecular determinants shaping the outcome of readthrough, namely the nucleotide and protein context, with the latter being pivotal to produce functional full-length proteins. Through the interpretation of experimental and mechanistic findings, mainly obtained in lysosomal and coagulation disorders, we also propose a scenario of potential readthrough-favorable features to achieve relevant rescue profiles, representing the main issue for the potential translatability of readthrough as a therapeutic strategy.
Keywords: nonsense mutations; premature termination codons; ribosome readthrough.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

