NOTCH1 Signaling in Head and Neck Squamous Cell Carcinoma
- PMID: 33322834
- PMCID: PMC7764697
- DOI: 10.3390/cells9122677
NOTCH1 Signaling in Head and Neck Squamous Cell Carcinoma
Abstract
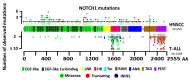
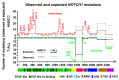
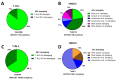
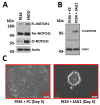
Biomarker-driven targeted therapies are lacking for head and neck squamous cell carcinoma (HNSCC), which is common and lethal. Efforts to develop such therapies are hindered by a genomic landscape dominated by the loss of tumor suppressor function, including NOTCH1 that is frequently mutated in HNSCC. Clearer understanding of NOTCH1 signaling in HNSCCs is crucial to clinically targeting this pathway. Structural characterization of NOTCH1 mutations in HNSCC demonstrates that most are predicted to cause loss of function, in agreement with NOTCH1's role as a tumor suppressor in this cancer. Experimental manipulation of NOTCH1 signaling in HNSCC cell lines harboring either mutant or wild-type NOTCH1 further supports a tumor suppressor function. Additionally, the loss of NOTCH1 signaling can drive HNSCC tumorigenesis and clinical aggressiveness. Our recent data suggest that NOTCH1 controls genes involved in early differentiation that could have different phenotypic consequences depending on the cancer's genetic background, including acquisition of pseudo-stem cell-like properties. The presence of NOTCH1 mutations may predict response to treatment with an immune checkpoint or phosphatidylinositol 3-kinase inhibitors. The latter is being tested in a clinical trial, and if validated, it may lead to the development of the first biomarker-driven targeted therapy for HNSCC.
Keywords: NOTCH1; head and neck squamous cell carcinoma; mutation; phosphatidylinositol 3-kinase; synthetic lethal; tumor suppressor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Chow L.Q.M., Haddad R., Gupta S., Mahipal A., Mehra R., Tahara M., Berger R., Eder J.P., Burtness B., Lee S.H., et al. Antitumor Activity of Pembrolizumab in Biomarker-Unselected Patients with Recurrent and/or Metastatic Head and Neck Squamous Cell Carcinoma: Results from the Phase Ib KEYNOTE-012 Expansion Cohort. J. Clin. Oncol. 2016;34:3838–3845. doi: 10.1200/JCO.2016.68.1478. - DOI - PMC - PubMed
-
- Bauml J., Seiwert T.Y., Pfister D.G., Worden F., Liu S.V., Gilbert J., Saba N.F., Weiss J., Wirth L., Sukari A., et al. Pembrolizumab for Platinum- and Cetuximab-Refractory Head and Neck Cancer: Results from a Single-Arm, Phase II Study. J. Clin. Oncol. 2017;35:1542–1549. doi: 10.1200/JCO.2016.70.1524. - DOI - PMC - PubMed
-
- Harrington K.J., Ferris R.L., Blumenschein G., Jr., Colevas A.D., Fayette J., Licitra L., Kasper S., Even C., Vokes E.E., Worden F., et al. Nivolumab versus standard, single-agent therapy of investigator’s choice in recurrent or metastatic squamous cell carcinoma of the head and neck (CheckMate 141): Health-related quality-of-life results from a randomised, phase 3 trial. Lancet Oncol. 2017;18:1104–1115. doi: 10.1016/S1470-2045(17)30421-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

