Hepatitis C reference viruses highlight potent antibody responses and diverse viral functional interactions with neutralising antibodies
- PMID: 33323394
- PMCID: PMC8355883
- DOI: 10.1136/gutjnl-2020-321190
Hepatitis C reference viruses highlight potent antibody responses and diverse viral functional interactions with neutralising antibodies
Erratum in
-
Correction: Hepatitis C reference viruses highlight potent antibody responses and diverse viral functional interactions with neutralising antibodies.Gut. 2024 Jan 5;73(2):e1. doi: 10.1136/gutjnl-2020-321190corr1. Gut. 2024. PMID: 38182159 No abstract available.
Abstract
Objective: Neutralising antibodies are key effectors of infection-induced and vaccine-induced immunity. Quantification of antibodies' breadth and potency is critical for understanding the mechanisms of protection and for prioritisation of vaccines. Here, we used a unique collection of human specimens and HCV strains to develop HCV reference viruses for quantification of neutralising antibodies, and to investigate viral functional diversity.
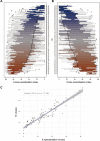
Design: We profiled neutralisation potency of polyclonal immunoglobulins from 104 patients infected with HCV genotype (GT) 1-6 across 13 HCV strains representing five viral GTs. Using metric multidimensional scaling, we plotted HCV neutralisation onto neutralisation maps. We employed K-means clustering to guide virus clustering and selecting representative strains.
Results: Viruses differed greatly in neutralisation sensitivity, with J6 (GT2a) being most resistant and SA13 (GT5a) being most sensitive. They mapped to six distinct neutralisation clusters, in part composed of viruses from different GTs. There was no correlation between viral neutralisation and genetic distance, indicating functional neutralisation clustering differs from sequence-based clustering. Calibrating reference viruses representing these clusters against purified antibodies from 496 patients infected by GT1 to GT6 viruses readily identified individuals with extraordinary potent and broadly neutralising antibodies. It revealed comparable antibody cross-neutralisation and diversity between specimens from diverse viral GTs, confirming well-balanced reporting of HCV cross-neutralisation across highly diverse human samples.
Conclusion: Representative isolates from six neutralisation clusters broadly reconstruct the functional HCV neutralisation space. They enable high resolution profiling of HCV neutralisation and they may reflect viral functional and antigenic properties important to consider in HCV vaccine design.
Keywords: HCV; genotype; hepatitis C; immunology in hepatology; liver.
© Author(s) (or their employer(s)) 2021. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: MC reports personal fees from Abbvie, personal fees from Gilead Sciences, personal fees from Janssen-Cilag, personal fees from Merck, MD, personal fees from Biogen, personal fees from Falk Foundation, personal fees from Siemens, personal fees from Spring Bank, personal fees from GlaxoSmithKline, outside the submitted work.
Figures






Comment in
-
Vaccines against hepatitis C: a travel into neutralisation space.Gut. 2021 Sep;70(9):1609-1610. doi: 10.1136/gutjnl-2020-323377. Epub 2021 Jan 25. Gut. 2021. PMID: 33495269 No abstract available.
References
-
- World Health Organisation . Global hepatitis report 2017. Geneva: WHO; 2017.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
