Merkel Cell Polyomavirus Encodes Circular RNAs (circRNAs) Enabling a Dynamic circRNA/microRNA/mRNA Regulatory Network
- PMID: 33323517
- PMCID: PMC7773998
- DOI: 10.1128/mBio.03059-20
Merkel Cell Polyomavirus Encodes Circular RNAs (circRNAs) Enabling a Dynamic circRNA/microRNA/mRNA Regulatory Network
Abstract
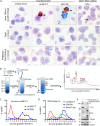
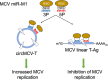
Viral noncoding RNAs have acquired increasing prominence as important regulators of infection and mediators of pathogenesis. Circular RNAs (circRNAs) generated by backsplicing events have been identified in several oncogenic human DNA viruses. Here, we show that Merkel cell polyomavirus (MCV), the etiologic cause of ∼80% of Merkel cell carcinomas (MCCs), also expresses circular RNAs. By RNase R-resistant RNA sequencing, four putative circRNA backsplice junctions (BSJs) were identified from the MCV early region (ER). The most abundantly expressed MCV circRNA, designated circMCV-T, is generated through backsplicing of all of ER exon II to form a 762-nucleotide (nt) circular RNA molecule. Curiously, circMCV-T, as well as two other less abundantly expressed putative MCV circRNAs, overlaps in a complementary fashion with the MCV microRNA (miRNA) locus that encodes MCV-miR-M1. circMCV-T is consistently detected in concert with linear T antigen transcripts throughout infection, suggesting a crucial role for this RNA molecule in the regulatory functions of the early region, known to be vital for viral replication. Knocking out the hairpin structure of MCV-miR-M1 in genomic early region expression constructs and using a new high-efficiency, recombinase-mediated, recircularized MCV molecular clone demonstrates that circMCV-T levels decrease in the presence of MCV-miR-M1, underscoring the interplay between MCV circRNA and miRNA. Furthermore, circMCV-T partially reverses the known inhibitory effect of MCV-miR-M1 on early gene expression. RNase R-resistant RNA sequencing of lytic rat polyomavirus 2 (RatPyV2) identified an analogously located circRNA, stipulating a crucial, conserved regulatory function of this class of RNA molecules in the family of polyomaviruses.IMPORTANCE Covalently closed circular RNAs were recently described in the human DNA tumor viruses Epstein-Barr virus (EBV), Kaposi's sarcoma-associated herpesvirus (KSHV), and human papillomavirus (HPV). Here, we show that MCV, another DNA tumor virus, generates circRNAs from its early regulatory region in concert with T antigen linear transcripts. MCV circMCV-T interacts with another MCV noncoding RNA, miR-M1, to functionally modulate early region transcript expression important for viral replication and long-term episomal persistence. This work describes a dynamic regulatory network integrating circRNA/miRNA/mRNA biomolecules and underscores the intricate functional modulation between several classes of polyomavirus-encoded RNAs in the control of viral replication.
Keywords: Merkel cell carcinoma; Merkel cell polyomavirus; T antigen; circular RNA; micro RNA; noncoding RNA; polyomaviruses.
Copyright © 2020 Abere et al.
Figures




References
-
- Theiss JM, Gunther T, Alawi M, Neumann F, Tessmer U, Fischer N, Grundhoff A. 2015. A comprehensive analysis of replicating Merkel cell polyomavirus genomes delineates the viral transcription program and suggests a role for mcv-miR-M1 in episomal persistence. PLoS Pathog 11:e1004974. doi:10.1371/journal.ppat.1004974. - DOI - PMC - PubMed
-
- Carter JJ, Daugherty MD, Qi X, Bheda-Malge A, Wipf GC, Robinson K, Roman A, Malik HS, Galloway DA. 2013. Identification of an overprinting gene in Merkel cell polyomavirus provides evolutionary insight into the birth of viral genes. Proc Natl Acad Sci U S A 110:12744–12749. doi:10.1073/pnas.1303526110. - DOI - PMC - PubMed
-
- Kwun HJ, Guastafierro A, Shuda M, Meinke G, Bohm A, Moore PS, Chang Y. 2009. The minimum replication origin of Merkel cell polyomavirus has a unique large T-antigen loading architecture and requires small T-antigen expression for optimal replication. J Virol 83:12118–12128. doi:10.1128/JVI.01336-09. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

