Characterization of the GABRB2-Associated Neurodevelopmental Disorders
- PMID: 33325057
- PMCID: PMC9161810
- DOI: 10.1002/ana.25985
Characterization of the GABRB2-Associated Neurodevelopmental Disorders
Erratum in
-
Correction.Ann Neurol. 2021 Nov;90(5):859. doi: 10.1002/ana.26215. Epub 2021 Oct 8. Ann Neurol. 2021. PMID: 34625987 No abstract available.
Abstract
Objective: We aimed to characterize the phenotypic spectrum and functional consequences associated with variants in the gene GABRB2, coding for the γ-aminobutyric acid type A (GABAA ) receptor subunit β2.
Methods: We recruited and systematically evaluated 25 individuals with variants in GABRB2, 17 of whom are newly described and 8 previously reported with additional clinical data. Functional analysis was performed using a Xenopus laevis oocyte model system.
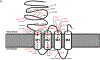
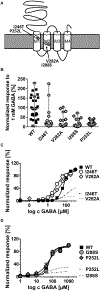
Results: Our cohort of 25 individuals from 22 families with variants in GABRB2 demonstrated a range of epilepsy phenotypes from genetic generalized epilepsy to developmental and epileptic encephalopathy. Fifty-eight percent of individuals had pharmacoresistant epilepsy; response to medications targeting the GABAergic pathway was inconsistent. Developmental disability (present in 84%) ranged from mild intellectual disability to severe global disability; movement disorders (present in 44%) included choreoathetosis, dystonia, and ataxia. Disease-associated variants cluster in the extracellular N-terminus and transmembrane domains 1-3, with more severe phenotypes seen in association with variants in transmembrane domains 1 and 2 and the allosteric binding site between transmembrane domains 2 and 3. Functional analysis of 4 variants in transmembrane domains 1 or 2 (p.Ile246Thr, p.Pro252Leu, p.Ile288Ser, p.Val282Ala) revealed strongly reduced amplitudes of GABA-evoked anionic currents.
Interpretation: GABRB2-related epilepsy ranges broadly in severity from genetic generalized epilepsy to developmental and epileptic encephalopathies. Developmental disability and movement disorder are key features. The phenotypic spectrum is comparable to other GABAA receptor-encoding genes. Phenotypic severity varies by protein domain. Experimental evidence supports loss of GABAergic inhibition as the mechanism underlying GABRB2-associated neurodevelopmental disorders. ANN NEUROL 2021;89:573-586.
© 2020 American Neurological Association.
Figures



References
-
- Baulac S, Huberfeld G, Gourfinkel-An I, Mitropoulou G, Beranger A, Prud’homme JF, et al. First genetic evidence of GABA(A) receptor dysfunction in epilepsy: a mutation in the gamma2-subunit gene. Nat Genet. 2001;28(1):46–8. - PubMed
-
- Hirose S Mutant GABA(A) receptor subunits in genetic (idiopathic) epilepsy. Prog Brain Res. 2014;213:55–85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

