High-throughput genotyping assays for identification of glycophorin B deletion variants in population studies
- PMID: 33325748
- PMCID: PMC8022085
- DOI: 10.1177/1535370220968545
High-throughput genotyping assays for identification of glycophorin B deletion variants in population studies
Abstract
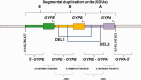
Glycophorins are the most abundant sialoglycoproteins on the surface of human erythrocyte membranes. Genetic variation in glycophorin region of human chromosome 4 (containing GYPA, GYPB, and GYPE genes) is of interest because the gene products serve as receptors for pathogens of major public health interest, including Plasmodiumsp., Babesiasp., Influenza virus, Vibrio cholerae El Tor Hemolysin, and Escherichia coli. A large structural rearrangement and hybrid glycophorin variant, known as Dantu, which was identified in East African populations, has been linked with a 40% reduction in risk for severe malaria. Apart from Dantu, other large structural variants exist, with the most common being deletion of the whole GYPB gene and its surrounding region, resulting in multiple different deletion forms. In West Africa particularly, these deletions are estimated to account for between 5 and 15% of the variation in different populations, mostly attributed to the forms known as DEL1 and DEL2. Due to the lack of specific variant assays, little is known of the distribution of these variants. Here, we report a modification of a previous GYPB DEL1 assay and the development of a novel GYPB DEL2 assay as high-throughput PCR-RFLP assays, as well as the identification of the crossover/breakpoint for GYPB DEL2. Using 393 samples from three study sites in Ghana as well as samples from HapMap and 1000 G projects for validation, we show that our assays are sensitive and reliable for genotyping GYPB DEL1 and DEL2. To the best of our knowledge, this is the first report of such high-throughput genotyping assays by PCR-RFLP for identifying specific GYPB deletion types in populations. These assays will enable better identification of GYPB deletions for large genetic association studies and functional experiments to understand the role of this gene cluster region in susceptibility to malaria and other diseases.
Keywords: GYPB deletion; Glycophorins; Plasmodium; invasion; malaria; red blood cell.
Conflict of interest statement
Figures





References
-
- WHO. World malaria report 2018. Luxembourg: World Health Organizaiton, 2018, p. 210
-
- Cowman AF, Crabb BS. Invasion of red blood cells by malaria parasites. Cell 2006; 124:755–66 - PubMed
-
- Gaur D, Mayer DG, Miller LH. Parasite ligand–host receptor interactions during invasion of erythrocytes by plasmodium merozoites. Int J Parasitol 2004; 34:1413–29 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

