Assessing the Role of Rare Genetic Variation in Patients With Heart Failure
- PMID: 33326012
- PMCID: PMC7745141
- DOI: 10.1001/jamacardio.2020.6500
Assessing the Role of Rare Genetic Variation in Patients With Heart Failure
Abstract
Importance: Sequencing studies have identified causal genetic variants for distinct subtypes of heart failure (HF) such as hypertrophic or dilated cardiomyopathy. However, the role of rare, high-impact variants in HF, for which ischemic heart disease is the leading cause, has not been systematically investigated.
Objective: To assess the contribution of rare variants to all-cause HF with and without reduced left ventricular ejection fraction.
Design, setting, and participants: This was a retrospective analysis of clinical trials and a prospective epidemiological resource (UK Biobank). Whole-exome sequencing of patients with HF was conducted from the Candesartan in Heart Failure-Assessment of Reduction in Mortality and Morbidity (CHARM) and Controlled Rosuvastatin Multinational Trial in Heart Failure (CORONA) clinical trials. Data were collected from March 1999 to May 2003 for the CHARM studies and September 2003 to July 2007 for the CORONA study. Using a gene-based collapsing approach, the proportion of patients with HF and controls carrying rare and presumed deleterious variants was compared. The burden of pathogenic variants in known cardiomyopathy genes was also investigated to assess the diagnostic yield. Exome sequencing data were generated between January 2018 and October 2018, and analysis began October 2018 and ended April 2020.
Main outcomes and measures: Odds ratios and P values for genes enriched for rare and presumed deleterious variants in either patients with HF or controls and diagnostic yield of pathogenic variants in known cardiomyopathy genes.
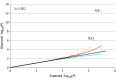
Results: This study included 5942 patients with HF and 13 156 controls. The mean (SD) age was 68.9 (9.9) years and 4213 (70.9%) were male. A significant enrichment of protein-truncating variants in the TTN gene (P = 3.35 × 10-13; odds ratio, 2.54; 95% CI, 1.96-3.31) that was further increased after restriction to variants in exons constitutively expressed in the heart (odds ratio, 4.52; 95% CI, 3.10-6.68). Validation using UK Biobank data showed a similar enrichment (odds ratio, 4.97; 95% CI, 3.94-6.19 after restriction). In the clinical trials, 201 of 5916 patients with HF (3.4%) had a pathogenic or likely pathogenic cardiomyopathy variant implicating 21 different genes. Notably, 121 of 201 individuals (60.2%) had ischemic heart disease as the clinically identified etiology for the HF. Individuals with HF and preserved ejection fraction had only a slightly lower yield than individuals with midrange or reduced ejection fraction (20 of 767 [2.6%] vs 15 of 392 [3.8%] vs 166 of 4757 [3.5%]).
Conclusions and relevance: An increased burden of diagnostic mendelian cardiomyopathy variants in a broad group of patients with HF of mostly ischemic etiology compared with controls was observed. This work provides further evidence that mendelian genetic conditions may represent an important subset of complex late-onset diseases such as HF, irrespective of the clinical presentation.
Conflict of interest statement
Figures

Comment in
-
Genetic Contribution to Common Heart Failure-Not So Rare?JAMA Cardiol. 2021 Apr 1;6(4):387. doi: 10.1001/jamacardio.2020.6508. JAMA Cardiol. 2021. PMID: 33325982 No abstract available.
References
-
- Vos T, Allen C, Arora M, et al. ; GBD 2015 Disease and Injury Incidence and Prevalence Collaborators . Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388(10053):1545-1602. doi:10.1016/S0140-6736(16)31678-6 - DOI - PMC - PubMed
-
- Yancy CW, Jessup M, Bozkurt B, et al. ; American College of Cardiology Foundation; American Heart Association Task Force on Practice Guidelines . 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2013;62(16):e147-e239. doi:10.1016/j.jacc.2013.05.019 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

