CD4+ follicular regulatory T cells optimize the influenza virus-specific B cell response
- PMID: 33326020
- PMCID: PMC7748821
- DOI: 10.1084/jem.20200547
CD4+ follicular regulatory T cells optimize the influenza virus-specific B cell response
Abstract
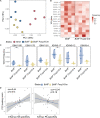
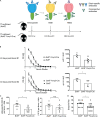
CD4+ follicular regulatory T (Tfr) cells control B cell responses through the modulation of follicular helper T (Tfh) cells and germinal center development while suppressing autoreactivity; however, their role in the regulation of productive germinal center B cell responses and humoral memory is incompletely defined. We show that Tfr cells promote antigen-specific germinal center B cell responses upon influenza virus infection. Following viral challenge, we found that Tfr cells are necessary for robust generation of virus-specific, long-lived plasma cells, antibody production against both hemagglutinin (HA) and neuraminidase (NA), the two major influenza virus glycoproteins, and appropriate regulation of the BCR repertoire. To further investigate the functional relevance of Tfr cells during viral challenge, we used a sequential immunization model with repeated exposure of antigenically partially conserved strains of influenza viruses, revealing that Tfr cells promote recall antibody responses against the conserved HA stalk region. Thus, Tfr cells promote antigen-specific B cell responses and are essential for the development of long-term humoral memory.
© 2020 Lu et al.
Conflict of interest statement
Disclosures: K.C. O’Connor reported personal fees from Alexion and grants from Ra Pharma outside the submitted work; is the recipient of a sponsored research subaward from the University of Pennsylvania, the primary financial sponsor of which is Cabaletta Bio; and is a consultant and equity shareholder of Cabaletta Bio. No other disclosures were reported.
Figures




References
-
- Botta, D., Fuller M.J., Marquez-Lago T.T., Bachus H., Bradley J.E., Weinmann A.S., Zajac A.J., Randall T.D., Lund F.E., León B., and Ballesteros-Tato A.. 2017. Dynamic regulation of T follicular regulatory cell responses by interleukin 2 during influenza infection. Nat. Immunol. 18:1249–1260. 10.1038/ni.3837 - DOI - PMC - PubMed
-
- Böttcher-Friebertshäuser, E., Freuer C., Sielaff F., Schmidt S., Eickmann M., Uhlendorff J., Steinmetzer T., Klenk H.-D., and Garten W.. 2010. Cleavage of influenza virus hemagglutinin by airway proteases TMPRSS2 and HAT differs in subcellular localization and susceptibility to protease inhibitors. J. Virol. 84:5605–5614. 10.1128/JVI.00140-10 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

