mRNA induced expression of human angiotensin-converting enzyme 2 in mice for the study of the adaptive immune response to severe acute respiratory syndrome coronavirus 2
- PMID: 33326500
- PMCID: PMC7773324
- DOI: 10.1371/journal.ppat.1009163
mRNA induced expression of human angiotensin-converting enzyme 2 in mice for the study of the adaptive immune response to severe acute respiratory syndrome coronavirus 2
Abstract
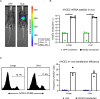
The novel human coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a pandemic. Critical to the rapid evaluation of vaccines and antivirals against SARS-CoV-2 is the development of tractable animal models to understand the adaptive immune response to the virus. To this end, the use of common laboratory strains of mice is hindered by significant divergence of the angiotensin-converting enzyme 2 (ACE2), which is the receptor required for entry of SARS-CoV-2. In the current study, we designed and utilized an mRNA-based transfection system to induce expression of the hACE2 receptor in order to confer entry of SARS-CoV-2 in otherwise non-permissive cells. By employing this expression system in an in vivo setting, we were able to interrogate the adaptive immune response to SARS-CoV-2 in type 1 interferon receptor deficient mice. In doing so, we showed that the T cell response to SARS-CoV-2 is enhanced when hACE2 is expressed during infection. Moreover, we demonstrated that these responses are preserved in memory and are boosted upon secondary infection. Importantly, using this system, we functionally identified the CD4+ and CD8+ structural peptide epitopes targeted during SARS-CoV-2 infection in H2b restricted mice and confirmed their existence in an established model of SARS-CoV-2 pathogenesis. We demonstrated that, identical to what has been seen in humans, the antigen-specific CD8+ T cells in mice primarily target peptides of the spike and membrane proteins, while the antigen-specific CD4+ T cells target peptides of the nucleocapsid, membrane, and spike proteins. As the focus of the immune response in mice is highly similar to that of the humans, the identification of functional murine SARS-CoV-2-specific T cell epitopes provided in this study will be critical for evaluation of vaccine efficacy in murine models of SARS-CoV-2 infection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Update of
-
mRNA induced expression of human angiotensin-converting enzyme 2 in mice for the study of the adaptive immune response to severe acute respiratory syndrome coronavirus 2.bioRxiv [Preprint]. 2020 Aug 7:2020.08.07.241877. doi: 10.1101/2020.08.07.241877. bioRxiv. 2020. Update in: PLoS Pathog. 2020 Dec 16;16(12):e1009163. doi: 10.1371/journal.ppat.1009163. PMID: 32793909 Free PMC article. Updated. Preprint.
Similar articles
-
mRNA induced expression of human angiotensin-converting enzyme 2 in mice for the study of the adaptive immune response to severe acute respiratory syndrome coronavirus 2.bioRxiv [Preprint]. 2020 Aug 7:2020.08.07.241877. doi: 10.1101/2020.08.07.241877. bioRxiv. 2020. Update in: PLoS Pathog. 2020 Dec 16;16(12):e1009163. doi: 10.1371/journal.ppat.1009163. PMID: 32793909 Free PMC article. Updated. Preprint.
-
Mapping and role of T cell response in SARS-CoV-2-infected mice.J Exp Med. 2021 Apr 5;218(4):e20202187. doi: 10.1084/jem.20202187. J Exp Med. 2021. PMID: 33464307 Free PMC article.
-
Modeling SARS-CoV-2 Infection in Mice Using Lentiviral hACE2 Vectors Infers Two Modes of Immune Responses to SARS-CoV-2 Infection.Viruses. 2021 Dec 21;14(1):11. doi: 10.3390/v14010011. Viruses. 2021. PMID: 35062215 Free PMC article.
-
Immunological Aspects of SARS-CoV-2 Infection and the Putative Beneficial Role of Vitamin-D.Int J Mol Sci. 2021 May 16;22(10):5251. doi: 10.3390/ijms22105251. Int J Mol Sci. 2021. PMID: 34065735 Free PMC article. Review.
-
The expression of hACE2 receptor protein and its involvement in SARS-CoV-2 entry, pathogenesis, and its application as potential therapeutic target.Tumour Biol. 2021;43(1):177-196. doi: 10.3233/TUB-200084. Tumour Biol. 2021. PMID: 34420993 Review.
Cited by
-
Production of SARS-CoV-2 virus-like particles as a vaccine candidate in stable cell lines through inducible E and M protein expression.BMC Biotechnol. 2025 Jul 28;25(1):79. doi: 10.1186/s12896-025-01016-x. BMC Biotechnol. 2025. PMID: 40717071 Free PMC article.
-
The role of vaccination route with an adenovirus-vectored vaccine in protection, viral control, and transmission in the SARS-CoV-2/K18-hACE2 mouse infection model.Front Immunol. 2023 Aug 16;14:1188392. doi: 10.3389/fimmu.2023.1188392. eCollection 2023. Front Immunol. 2023. PMID: 37662899 Free PMC article.
-
The Serological Sciences Network (SeroNet) for COVID-19: Depth and Breadth of Serology Assays and Plans for Assay Harmonization.medRxiv [Preprint]. 2022 Mar 14:2022.02.27.22271399. doi: 10.1101/2022.02.27.22271399. medRxiv. 2022. Update in: mSphere. 2022 Aug 31;7(4):e0019322. doi: 10.1128/msphere.00193-22. PMID: 35262095 Free PMC article. Updated. Preprint.
-
MCMV-based vaccine vectors expressing full-length viral proteins provide long-term humoral immune protection upon a single-shot vaccination.Cell Mol Immunol. 2022 Feb;19(2):234-244. doi: 10.1038/s41423-021-00814-5. Epub 2022 Jan 7. Cell Mol Immunol. 2022. PMID: 34992275 Free PMC article.
-
Post-acute immunological and behavioral sequelae in mice after Omicron infection.bioRxiv [Preprint]. 2023 Oct 4:2023.06.05.543758. doi: 10.1101/2023.06.05.543758. bioRxiv. 2023. PMID: 37333294 Free PMC article. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

