Evidence For Long-Lasting Transgenerational Antiviral Immunity in Insects
- PMID: 33326778
- PMCID: PMC7758158
- DOI: 10.1016/j.celrep.2020.108506
Evidence For Long-Lasting Transgenerational Antiviral Immunity in Insects
Abstract
Transgenerational immune priming (TGIP) allows memory-like immune responses to be transmitted from parents to offspring in many invertebrates. Despite increasing evidence for TGIP in insects, the mechanisms involved in the transfer of information remain largely unknown. Here, we show that Drosophila melanogaster and Aedes aegypti transmit antiviral immunological memory to their progeny that lasts throughout generations. We observe that TGIP, which is virus and sequence specific but RNAi independent, is initiated by a single exposure to disparate RNA viruses and also by inoculation of a fragment of viral double-stranded RNA. The progeny, which inherit a viral DNA that is only a fragment of the viral RNA used to infect the parents, display enriched expression of genes related to chromatin and DNA binding. These findings represent a demonstration of TGIP for RNA viruses in invertebrates, broadly increasing our understanding of the immune response, host genome plasticity, and antiviral memory of the germline.
Keywords: Aedes aegypti; Drosophila melanogaster; RNA viruses; adaptive immunity; antiviral memory; chromatin modifications; immune response; insects; transgenerational immune priming.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures
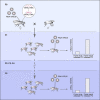


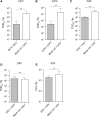

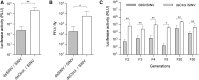

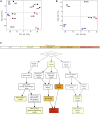
References
-
- Contreras-Garduño J., Lanz-Mendoza H., Franco B., Nava A., Pedraza-Reyes M., Canales-Lazcano J. Insect immune priming: ecology and experimental evidences. Ecol. Entomol. 2016;41:351–366.
-
- Dhinaut J., Chogne M., Moret Y. Immune priming specificity within and across generations reveals the range of pathogens affecting evolution of immunity in an insect. J. Anim. Ecol. 2018;87:448–463. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

