An ACE2 Microbody Containing a Single Immunoglobulin Fc Domain Is a Potent Inhibitor of SARS-CoV-2
- PMID: 33326798
- PMCID: PMC7705358
- DOI: 10.1016/j.celrep.2020.108528
An ACE2 Microbody Containing a Single Immunoglobulin Fc Domain Is a Potent Inhibitor of SARS-CoV-2
Abstract
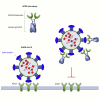
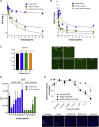
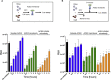
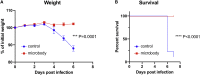
Soluble forms of angiotensin-converting enzyme 2 (ACE2) have recently been shown to inhibit severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. We report on an improved soluble ACE2, termed a "microbody," in which the ACE2 ectodomain is fused to Fc domain 3 of the immunoglobulin (Ig) heavy chain. The protein is smaller than previously described ACE2-Ig Fc fusion proteins and contains an H345A mutation in the ACE2 catalytic active site that inactivates the enzyme without reducing its affinity for the SARS-CoV-2 spike. The disulfide-bonded ACE2 microbody protein inhibits entry of SARS-CoV-2 spike protein pseudotyped virus and replication of live SARS-CoV-2 in vitro and in a mouse model. Its potency is 10-fold higher than soluble ACE2, and it can act after virus bound to the cell. The microbody inhibits the entry of β coronaviruses and virus with the variant D614G spike. The ACE2 microbody may be a valuable therapeutic for coronavirus disease 2019 (COVID-19) that is active against viral variants and future coronaviruses.
Keywords: ACE2 transgenic; D614G; Fc fusion; SARS-CoV-2; coronavirus; entry inhibitor; lentiviral pseudotype; microbody; soluble ACE2; spike protein.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests A provisional patent application has been filed with the US Patent and Trademark Office.
Figures




References
-
- Case J.B., Rothlauf P.W., Chen R.E., Liu Z., Zhao H., Kim A.S., Bloyet L.M., Zeng Q., Tahan S., Droit L. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.05.18.102038. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

