Genomic Epidemiology of the First Wave of SARS-CoV-2 in Italy
- PMID: 33327566
- PMCID: PMC7765063
- DOI: 10.3390/v12121438
Genomic Epidemiology of the First Wave of SARS-CoV-2 in Italy
Abstract
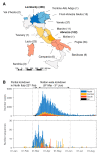
Italy was one of the first countries to experience a major epidemic of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), with >1000 cases confirmed by 1 March 2020. However, virus genome sequence data is sparse and there has been only limited investigation of virus transmission across the country. Here, we provide the most extensive study to date of the genomic epidemiology of SARS-CoV-2 in Italy covering the first wave of infection. We generated 191 new full-length genomes, largely sampled from central Italy (Abruzzo), before, during, and after the enforcement of a nationwide "lockdown" (8 March-3 June). These were combined with 460 published SARS-CoV-2 sequences sampled across Italy. Phylogenetic analysis including global sequence data revealed multiple independent introductions into Italy, with at least 124 instances of sequence clusters representing longer chains of transmission. Eighteen of these transmission clusters emerged before the nation-wide lockdown was implemented on 8 March, and an additional 18 had evidence for transmission between different Italian regions. Extended transmission periods between infections of up to 104 days were observed in five clusters. In addition, we found seven clusters that persisted throughout the lockdown period. Overall, we show how importations were an important driver of the first wave of SARS-CoV-2 in Italy.
Keywords: Italy; SARS-Cov-2; lockdown; phylogeny; transmission.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

