Identification of histone acetylation in a murine model of allergic asthma by proteomic analysis
- PMID: 33327783
- PMCID: PMC8024510
- DOI: 10.1177/1535370220980345
Identification of histone acetylation in a murine model of allergic asthma by proteomic analysis
Abstract
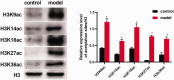
The pathogenesis of asthma is closely related to histone acetylation modification, but the specific acetylation sites related to this process remain indistinct. Herein, our study sought to identify differentially modified acetylation sites and their expression distribution in cells involved in asthma in lung tissues. The airway hyper-responsiveness, inflammation, and remodeling were assessed by non-invasive whole-body plethysmography, ELISA, and hematoxylin-eosin staining to confirm the successful establishment of the allergic asthma model. Afterward, the differentially modified acetylation sites in asthmatic lung tissues were identified and validated by using proteomics and western blotting, respectively. The immunohistochemistry analysis was applied to reveal the distribution of identified acetylation sites in asthmatic lung tissues. A total of 15 differentially modified acetylation sites, including 13 upregulated (H3K9ac, H3K14ac, H3K18ac, H3K23ac,H3K27ac, H3K36ac, H2B1KK120ac, H2B2BK20ac, H2BK16ac, H2BK20ac, H2BK108ac, H2BK116ac, and H2BK120ac) and 2 downregulated (H2BK5ac and H2BK11ac) sites were identified and validated. Furthermore, immunohistochemical staining of lung tissues showed that nine of the identified histone acetylation sites (H2BK5, H2BK11, H3K18, H2BK116, H2BK20, H2BK120, H3K9, H3K36, and H3K27) were differentially expressed in airway epithelial cells, and the acetylation of identified H3 histones were observed in both eosinophil and perivascular inflammatory cells. Additionally, differential expression of histone acetylation sites was also observed in nucleus of airway epithelial cells, vascular smooth muscle cells, perivascular inflammatory cells, and airway smooth muscle cells. In conclusion, we identified potential acetylation sites associated with asthma pathogenesis. These findings may contribute greatly in the search for therapeutic approaches for allergic asthma.
Keywords: Bronchial asthma; histone acetylation; proteomics; tandem mass tag.
Conflict of interest statement
Figures





References
-
- Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, Heinzmann A, Simma B, Frischer T, Willis-Owen SA, Wong KC, Illig T, Vogelberg C, Weiland SK, von Mutius E, Abecasis GR, Farrall M, Gut IG, Lathrop GM, Cookson WO. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 2007; 448:470–3 - PubMed
-
- Graff J, Tsai LH. Histone acetylation: molecular mnemonics on the chromatin. Nat Rev Neurosci 2013; 14:97–111 - PubMed
-
- Kidd CD, Thompson PJ, Barrett L, Baltic S. Histone modifications and asthma. The interface of the epigenetic and genetic landscapes. Am J Respir Cell Mol Biol 2016; 54:3–12 - PubMed

