Genomic and phenotypic characterisation of invasive neonatal and colonising group B Streptococcus isolates from Slovenia, 2001-2018
- PMID: 33327946
- PMCID: PMC7739447
- DOI: 10.1186/s12879-020-05599-y
Genomic and phenotypic characterisation of invasive neonatal and colonising group B Streptococcus isolates from Slovenia, 2001-2018
Abstract
Background: Group B Streptococcus (GBS) is the leading cause of invasive neonatal disease in the industrialized world. We aimed to genomically and phenotypically characterise invasive GBS isolates in Slovenia from 2001 to 2018 and contemporary colonising GBS isolates from screening cultures in 2018.
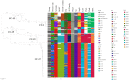
Methods: GBS isolates from 101 patients (invasive isolates) and 70 pregnant women (colonising isolates) were analysed. Basic clinical characteristics of the patients were collected from medical records. Antimicrobial susceptibility and phenotypic capsular serotype were determined. Whole-genome sequencing was performed to assign multilocus sequence types (STs), clonal complexes (CCs), pathogenicity/virulence factors, including capsular genotypes, and genome-based phylogeny.
Results: Among invasive neonatal disease patients, 42.6% (n = 43) were females, 41.5% (n = 39/94) were from preterm deliveries (< 37 weeks gestation), and 41.6% (n = 42) had early-onset disease (EOD). All isolates were susceptible to benzylpenicillin with low minimum inhibitory concentrations (MICs; ≤0.125 mg/L). Overall, 7 serotypes were identified (Ia, Ib, II-V and VIII); serotype III being the most prevalent (59.6%). Twenty-eight MLST STs were detected that clustered into 6 CCs. CC-17 was the most common CC overall (53.2%), as well as among invasive (67.3%) and non-invasive (32.9%) isolates (p < 0.001). CC-17 was more common among patients with late-onset disease (LOD) (81.4%) compared to EOD (47.6%) (p < 0.001). The prevalence of other CCs was 12.9% (CC-23), 11.1% (CC-12), 10.5% (CC-1), 8.2% (CC-19), and 1.8% (CC-498). Of all isolates, 2.3% were singletons.
Conclusions: A high prevalence of hypervirulent CC-17 isolates, with low genomic diversity and characteristic profile of pathogenicity/virulence factors, was detected among invasive neonatal and colonising GBS isolates from pregnant women in Slovenia. This is the first genomic characterisation of GBS isolates in Slovenia and provides valuable microbiological and genomic baseline data regarding the invasive and colonising GBS population nationally. Continuous genomic surveillance of GBS infections is crucial to analyse the impact of IND prevention strategies on the population structure of GBS locally, nationally, and internationally.
Keywords: Capsular type; GBS; Group B Streptococcus; Hypervirulent CC-17; Molecular epidemiology; Neonatal infection; Pathogenicity/virulence factors; Slovenia.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Verani JR, McGee L, Schrag SJ. Division of Bacterial Diseases, National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention (CDC). Prevention of perinatal group B streptococcal disease--revised guidelines from CDC, 2010. MMWR Recomm Rep. 2010;59:1–36. - PubMed
-
- Lasič M, Lučovnik M, Kaparič T, Ciringer M, Krivec JL, Fister P, et al. Invazivne okužbe novorojenčkov z bakterijo Streptococcus agalactiae v Sloveniji, 2003–2013. Zdrav Vestn. 2017;86:493–506.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

