Data structures based on k-mers for querying large collections of sequencing data sets
- PMID: 33328168
- PMCID: PMC7849385
- DOI: 10.1101/gr.260604.119
Data structures based on k-mers for querying large collections of sequencing data sets
Abstract
High-throughput sequencing data sets are usually deposited in public repositories (e.g., the European Nucleotide Archive) to ensure reproducibility. As the amount of data has reached petabyte scale, repositories do not allow one to perform online sequence searches, yet, such a feature would be highly useful to investigators. Toward this goal, in the last few years several computational approaches have been introduced to index and query large collections of data sets. Here, we propose an accessible survey of these approaches, which are generally based on representing data sets as sets of k-mers. We review their properties, introduce a classification, and present their general intuition. We summarize their performance and highlight their current strengths and limitations.
© 2021 Marchet et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
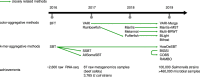
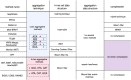
References
-
- Almodaresi F, Pandey P, Patro R. 2017. Rainbowfish: a succinct colored de Bruijn graph representation. In Proceedings of the Seventeenth International Workshop on Algorithms in Bioinformatics, Boston Dagstuhl Publishing, Saarbrücken/Wadern, Germany.
-
- Almodaresi F, Pandey P, Ferdman M, Johnson R, Patro R. 2019. An efficient, scalable and exact representation of high-dimensional color information enabled via de Bruijn graph search. In Proceedings of the International Conference on Research in Computational Molecular Biology, Washington, pp. 1–18. Springer, New York. - PMC - PubMed
-
- Bender MA, Farach-Colton M, Johnson R, Kraner R, Kuszmaul BC, Medjedovic D, Montes P, Shetty P, Spillane RP, Zadok E. 2012. Don't thrash: how to cache your hash on flash. PVLDB 5: 1627–1637.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
