Genome Number and Size Polymorphism in Zika Virus Infectious Units
- PMID: 33328311
- PMCID: PMC8094933
- DOI: 10.1128/JVI.00787-20
Genome Number and Size Polymorphism in Zika Virus Infectious Units
Abstract
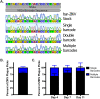
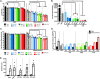
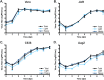
Zika virus (ZIKV; Flaviviridae, Flavivirus) is an arthropod-borne infection that can result in severe outcomes, particularly in fetuses infected in utero It has been assumed that infection by ZIKV, as well as other viruses, is largely initiated by individual virus particles binding to and entering a cell. However, recent studies have demonstrated that multiple virus particles are frequently delivered to a cell simultaneously and that this collective particle delivery enhances infection. ZIKV is maintained in nature between Aedes aegypti mosquitos and vertebrate hosts, including humans. Human infection is initiated through the injection of a relatively small initial inoculum comprised of a genetically complex virus population. Since most mutations decrease virus fitness, collective particle transmission could benefit ZIKV and other arthropod-borne diseases by facilitating the maintenance of genetic complexity and adaptability during infection or through other mechanisms. Therefore, we utilized a barcoded ZIKV to quantify the number of virus genomes that initiate a plaque. We found that individual plaques contain a mean of 10 infecting viral genomes (range, 1 to 212). Few plaques contained more than two dominant genomes. To determine whether multigenome infectious units consist of collectively transmitting virions, infectious units of ZIKV were then separated mechanically by centrifugation, and heavier fractions were found to contain more genomes per plaque-forming unit, with larger diameters. Finally, larger/heavier infectious units reformed after removal. These data suggest that ZIKV populations consist of a variety of infectious unit sizes, likely mostly made up of aggregates, and only rarely begin with a single virus genome.IMPORTANCE The arthropod-borne Zika virus (ZIKV) infects humans and can cause severe neurological sequelae, particularly in fetuses infected in utero How this virus has been able to spread across vast geological ranges and evolve in new host populations is not yet understood. This research demonstrates a novel mechanism of ZIKV transmission through multigenome aggregates, providing insight into ZIKV evolution, immunologic evasion, and better future therapeutic design. This study shows that ZIKV plaques result from collections of genomes rather than individual genomes, increasing the potential for interactions between ZIKV genotypes.
Keywords: ZIKV; Zika virus; aggregates; barcoded virus; flavivirus; specific infectivity.
Copyright © 2021 Sexton et al.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

