Chi3l1/YKL-40 is controlled by the astrocyte circadian clock and regulates neuroinflammation and Alzheimer's disease pathogenesis
- PMID: 33328329
- PMCID: PMC7808313
- DOI: 10.1126/scitranslmed.aax3519
Chi3l1/YKL-40 is controlled by the astrocyte circadian clock and regulates neuroinflammation and Alzheimer's disease pathogenesis
Abstract
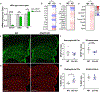
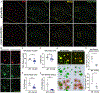
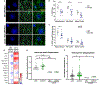
Regulation of glial activation and neuroinflammation are critical factors in the pathogenesis of Alzheimer's disease (AD). YKL-40, a primarily astrocytic protein encoded by the gene Chi3l1, is a widely studied cerebrospinal fluid biomarker that increases with aging and early in AD. However, the function of Chi3l1/YKL-40 in AD is unknown. In a cohort of patients with AD, we observed that a variant in the human CHI3L1 gene, which results in decreased CSF YKL-40 expression, was associated with slower AD progression. At baseline, Chi3l1 deletion in mice had no effect on astrocyte activation while modestly promoting microglial activation. In a mouse APP/PS1 model of AD, Chi3l1 deletion decreased amyloid plaque burden and increased periplaque expression of the microglial lysosomal marker CD68, suggesting that Chi3l1 may suppress glial phagocytic activation and promote amyloid accumulation. Accordingly, Chi3l1 knockdown increased phagocytosis of zymosan particles and of β-amyloid peptide in both astrocytes and microglia in vitro. We further observed that expression of Chi3l1 is regulated by the circadian clock, as deletion of the core clock proteins BMAL1 or CLOCK/NPAS2 strongly suppresses basal Chi3l1 expression, whereas deletion of the negative clock regulators PER1/PER2 increased Chi3l1 expression. Basal Chi3l1 mRNA was nonrhythmic because of a long mRNA half-life in astrocytes. However, inflammatory induction of Chi3l1 was gated by the clock. Our findings reveal Chi3l1/YKL-40 as a modulator of glial phagocytic activation and AD pathogenesis in both mice and humans and suggest that the astrocyte circadian clock regulates inflammatory Chi3l1 induction.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures




References
-
- Arranz AM, De Strooper B, The role of astroglia in Alzheimer’s disease: pathophysiology and clinical implications. Lancet Neurol, (2019). - PubMed
-
- Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, Bennett ML, Munch AE, Chung WS, Peterson TC, Wilton DK, Frouin A, Napier BA, Panicker N, Kumar M, Buckwalter MS, Rowitch DH, Dawson VL, Dawson TM, Stevens B, Barres BA, Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487 (2017). - PMC - PubMed
-
- Craig-Schapiro R, Perrin RJ, Roe CM, Xiong C, Carter D, Cairns NJ, Mintun MA, Peskind ER, Li G, Galasko DR, Clark CM, Quinn JF, D’Angelo G, Malone JP, Townsend RR, Morris JC, Fagan AM, Holtzman DM, YKL-40: a novel prognostic fluid biomarker for preclinical Alzheimer’s disease. Biological psychiatry 68, 903–912 (2010). - PMC - PubMed
-
- Sutphen CL, Jasielec MS, Shah AR, Macy EM, Xiong C, Vlassenko AG, Benzinger TL, Stoops EE, Vanderstichele HM, Brix B, Darby HD, Vandijck ML, Ladenson JH, Morris JC, Holtzman DM, Fagan AM, Longitudinal Cerebrospinal Fluid Biomarker Changes in Preclinical Alzheimer Disease During Middle Age. JAMA Neurol 72, 1029–1042 (2015). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG064877/AG/NIA NIH HHS/United States
- R01 AG058501/AG/NIA NIH HHS/United States
- RF1 AG044546/AG/NIA NIH HHS/United States
- R01 AG054517/AG/NIA NIH HHS/United States
- P30 AG066444/AG/NIA NIH HHS/United States
- U01 AG058922/AG/NIA NIH HHS/United States
- R01 AG044546/AG/NIA NIH HHS/United States
- U01 AG052411/AG/NIA NIH HHS/United States
- RF1 AG053303/AG/NIA NIH HHS/United States
- P01 AG003991/AG/NIA NIH HHS/United States
- P50 AG005681/AG/NIA NIH HHS/United States
- P01 AG026276/AG/NIA NIH HHS/United States
- RF1 AG058501/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

