Identifying cell-enriched miRNAs in kidney injury and repair
- PMID: 33328386
- PMCID: PMC7819746
- DOI: 10.1172/jci.insight.140399
Identifying cell-enriched miRNAs in kidney injury and repair
Abstract
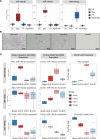
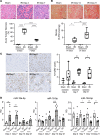
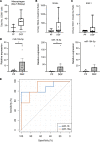
Small noncoding RNAs, miRNAs (miRNAs), are emerging as important modulators in the pathogenesis of kidney disease, with potential as biomarkers of kidney disease onset, progression, or therapeutic efficacy. Bulk tissue small RNA-sequencing (sRNA-Seq) and microarrays are widely used to identify dysregulated miRNA expression but are limited by the lack of precision regarding the cellular origin of the miRNA. In this study, we performed cell-specific sRNA-Seq on tubular cells, endothelial cells, PDGFR-β+ cells, and macrophages isolated from injured and repairing kidneys in the murine reversible unilateral ureteric obstruction model. We devised an unbiased bioinformatics pipeline to define the miRNA enrichment within these cell populations, constructing a miRNA catalog of injury and repair. Our analysis revealed that a significant proportion of cell-specific miRNAs in healthy animals were no longer specific following injury. We then applied this knowledge of the relative cell specificity of miRNAs to deconvolute bulk miRNA expression profiles in the renal cortex in murine models and human kidney disease. Finally, we used our data-driven approach to rationally select macrophage-enriched miR-16-5p and miR-18a-5p and demonstrate that they are promising urinary biomarkers of acute kidney injury in renal transplant recipients.
Keywords: Bioinformatics; Nephrology; Noncoding RNAs; Organ transplantation.
Conflict of interest statement
Figures





References
-
- Connor KL, Denby L. MicroRNAs as non-invasive biomarkers of renal disease [published online September 20, 2019]. Nephrol Dial Transplant. https://dx.doi.org10.1093/ndt/gfz183. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

