A single dose of recombinant VSV-∆G-spike vaccine provides protection against SARS-CoV-2 challenge
- PMID: 33328475
- PMCID: PMC7745033
- DOI: 10.1038/s41467-020-20228-7
A single dose of recombinant VSV-∆G-spike vaccine provides protection against SARS-CoV-2 challenge
Abstract
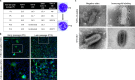
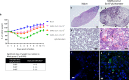
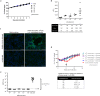
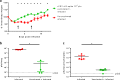
The COVID-19 pandemic caused by SARS-CoV-2 imposes an urgent need for rapid development of an efficient and cost-effective vaccine, suitable for mass immunization. Here, we show the development of a replication competent recombinant VSV-∆G-spike vaccine, in which the glycoprotein of VSV is replaced by the spike protein of SARS-CoV-2. In-vitro characterization of this vaccine indicates the expression and presentation of the spike protein on the viral membrane with antigenic similarity to SARS-CoV-2. A golden Syrian hamster in-vivo model for COVID-19 is implemented. We show that a single-dose vaccination results in a rapid and potent induction of SARS-CoV-2 neutralizing antibodies. Importantly, vaccination protects hamsters against SARS-CoV-2 challenge, as demonstrated by the abrogation of body weight loss, and alleviation of the extensive tissue damage and viral loads in lungs and nasal turbinates. Taken together, we suggest the recombinant VSV-∆G-spike as a safe, efficacious and protective vaccine against SARS-CoV-2.
Conflict of interest statement
Patent application for the described vaccine was filed by the Israel Institute for Biological Research.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

