RNAi-Mediated Suppression of Laccase2 Impairs Cuticle Tanning and Molting in the Cotton Boll Weevil (Anthonomus grandis)
- PMID: 33329040
- PMCID: PMC7717984
- DOI: 10.3389/fphys.2020.591569
RNAi-Mediated Suppression of Laccase2 Impairs Cuticle Tanning and Molting in the Cotton Boll Weevil (Anthonomus grandis)
Abstract
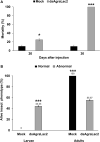
The cotton boll weevil, Anthonomus grandis, is the most economically important pest of cotton in Brazil. Pest management programs focused on A. grandis are based mostly on the use of chemical insecticides, which may cause serious ecological impacts. Furthermore, A. grandis has developed resistance to some insecticides after their long-term use. Therefore, alternative control approaches that are more sustainable and have reduced environmental impacts are highly desirable to protect cotton crops from this destructive pest. RNA interference (RNAi) is a valuable reverse genetics tool for the investigation of gene function and has been explored for the development of strategies to control agricultural insect pests. This study aimed to evaluate the biological role of the Laccase2 (AgraLac2) gene in A. grandis and its potential as an RNAi target for the control of this insect pest. We found that AgraLac2 is expressed throughout the development of A. grandis with significantly higher expression in pupal and adult developmental stages. In addition, the immunolocalization of the AgraLac2 protein in third-instar larvae using specific antibodies revealed that AgraLac2 is distributed throughout the epithelial tissue, the cuticle and the tracheal system. We also verified that the knockdown of AgraLac2 in A. grandis resulted in an altered cuticle tanning process, molting defects and arrested development. Remarkably, insects injected with dsAgraLac2 exhibited defects in cuticle hardening and pigmentation. As a consequence, the development of dsAgraLac2-treated insects was compromised, and in cases of severe phenotypic defects, the insects subsequently died. On the contrary, insects subjected to control treatments did not show any visible phenotypic defects in cuticle formation and successfully molted to the pupal and adult stages. Taken together, our data indicate that AgraLac2 is involved in the cuticle tanning process in A. grandis and may be a promising target for the development of RNAi-based technologies.
Keywords: RNAi; Laccase2; cuticle tanning; gene silencing; insect pest control.
Copyright © 2020 Firmino, Pinheiro, Moreira-Pinto, Antonino, Macedo, Martins-de-Sa, Arraes, Coelho, Fonseca, Silva, Engler, Silva, Lourenço-Tessutti, Terra and Grossi-de-Sa.
Figures






References
-
- Andersen S. O. (2012). “Cuticular sclerotization and tanning,” in Insect Molecular Biology and Biochemistry, ed. Gilbert L. I. (Amsterdam: Elsevier; ), 167–192. 10.1016/B978-0-12-384747-8.10006-6 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

