Oncology and Pharmacogenomics Insights in Polycystic Ovary Syndrome: An Integrative Analysis
- PMID: 33329391
- PMCID: PMC7729301
- DOI: 10.3389/fendo.2020.585130
Oncology and Pharmacogenomics Insights in Polycystic Ovary Syndrome: An Integrative Analysis
Abstract
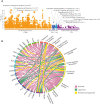
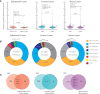
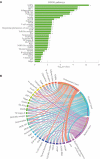
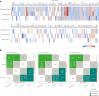
Polycystic ovary syndrome (PCOS) is a heterogeneous endocrine disorder characterized by hyperandrogenism, ovulatory dysfunction, and polycystic ovaries. Epidemiological findings revealed that women with PCOS are prone to develop certain cancer types due to their shared metabolic and endocrine abnormalities. However, the mechanism that relates PCOS and oncogenesis has not been addressed. Herein, in this review article the genomic status, transcriptional and protein profiles of 264 strongly PCOS related genes (PRG) were evaluated in endometrial cancer (EC), ovarian cancer (OV) and breast cancer (BC) exploring oncogenic databases. The genomic alterations of PRG were significantly higher when compared with a set of non-diseases genes in all cancer types. PTEN had the highest number of mutations in EC, TP53, in OC, and FSHR, in BC. Based on clinical data, women older than 50 years and Black or African American females carried the highest ratio of genomic alterations among all cancer types. The most altered signaling pathways were p53 in EC and OC, while Fc epsilon RI in BC. After evaluating PRG in normal and cancer tissue, downregulation of the differentially expressed genes was a common feature. Less than 30 proteins were up and downregulated in all cancer contexts. We identified 36 highly altered genes, among them 10 were shared between the three cancer types analyzed, which are involved in the cell proliferation regulation, response to hormone and to endogenous stimulus. Despite limited PCOS pharmacogenomics studies, 10 SNPs are reported to be associated with drug response. All were missense mutations, except for rs8111699, an intronic variant characterized as a regulatory element and presumably binding site for transcription factors. In conclusion, in silico analysis revealed key genes that might participate in PCOS and oncogenesis, which could aid in early cancer diagnosis. Pharmacogenomics efforts have implicated SNPs in drug response, yet still remain to be found.
Keywords: bioinformatic; breast cancer (BC); endometrial cancer (EC); ovarian cancer (OC); pharmacogenomics; polycystic ovary syndrome (PCOS).
Copyright © 2020 Yumiceba, López-Cortés, Pérez-Villa, Yumiseba, Guerrero, García-Cárdenas, Armendáriz-Castillo, Guevara-Ramírez, Leone, Zambrano and Paz-y-Miño.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

