Lipopolysaccharide Recognition in the Crossroads of TLR4 and Caspase-4/11 Mediated Inflammatory Pathways
- PMID: 33329561
- PMCID: PMC7732686
- DOI: 10.3389/fimmu.2020.585146
Lipopolysaccharide Recognition in the Crossroads of TLR4 and Caspase-4/11 Mediated Inflammatory Pathways
Erratum in
-
Corrigendum: Lipopolysaccharide Recognition in the Crossroads of TLR4 and Caspase-4/11 Mediated Inflammatory Pathways.Front Immunol. 2021 Jan 28;12:649442. doi: 10.3389/fimmu.2021.649442. eCollection 2021. Front Immunol. 2021. PMID: 33584736 Free PMC article.
Abstract
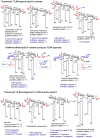
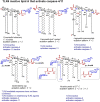
The innate immune response to lipopolysaccharide is essential for host defense against Gram-negative bacteria. In response to bacterial infection, the TLR4/MD-2 complex that is expressed on the surface of macrophages, monocytes, dendritic, and epithelial cells senses picomolar concentrations of endotoxic LPS and triggers the production of various pro-inflammatory mediators. In addition, LPS from extracellular bacteria which is either endocytosed or transfected into the cytosol of host cells or cytosolic LPS produced by intracellular bacteria is recognized by cytosolic proteases caspase-4/11 and hosts guanylate binding proteins that are involved in the assembly and activation of the NLRP3 inflammasome. All these events result in the initiation of pro-inflammatory signaling cascades directed at bacterial eradication. However, TLR4-mediated signaling and caspase-4/11-induced pyroptosis are largely involved in the pathogenesis of chronic and acute inflammation. Both extra- and intracellular LPS receptors-TLR4/MD-2 complex and caspase-4/11, respectively-are able to directly bind the lipid A motif of LPS. Whereas the structural basis of lipid A recognition by the TLR4 complex is profoundly studied and well understood, the atomic mechanism of LPS/lipid A interaction with caspase-4/11 is largely unknown. Here we describe the LPS-induced TLR4 and caspase-4/11 mediated signaling pathways and their cross-talk and scrutinize specific structural features of the lipid A motif of diverse LPS variants that have been reported to activate caspase-4/11 or to induce caspase-4/11 mediated activation of NLRP3 inflammasome (either upon transfection of LPS in vitro or upon infection of cell cultures with intracellular bacteria or by LPS as a component of the outer membrane vesicles). Generally, inflammatory caspases show rather similar structural requirements as the TLR4/MD-2 complex, so that a "basic" hexaacylated bisphosphorylated lipid A architecture is sufficient for activation. However, caspase-4/11 can sense and respond to much broader variety of lipid A variants compared to the very "narrow" specificity of TLR4/MD-2 complex as far as the number and the length of lipid chains attached at the diglucosamine backbone of lipid A is concerned. Besides, modification of the lipid A phosphate groups with positively charged appendages such as phosphoethanolamine or aminoarabinose could be essential for the interaction of lipid A/LPS with inflammatory caspases and related proteins.
Keywords: TLR4/MD-2; aminoarabinose; chemical structure; inflammation; innate immunity; lipid A; molecular recognition; structural basis.
Copyright © 2020 Zamyatina and Heine.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

