Differential Expression and Bioinformatics Analysis of circRNA in Non-small Cell Lung Cancer
- PMID: 33329727
- PMCID: PMC7732606
- DOI: 10.3389/fgene.2020.586814
Differential Expression and Bioinformatics Analysis of circRNA in Non-small Cell Lung Cancer
Abstract
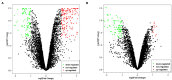
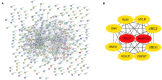
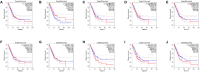
Circular RNA (CircRNA) plays an important role in tumorigenesis and progression of non-small cell lung cancer (NSCLC), but the pathogenesis of NSCLC caused by circRNA has not been fully elucidated. This study aimed to investigate differentially expressed circRNAs and identify the underlying pathogenesis hub genes of NSCLC by comprehensive bioinformatics analysis. Data of gene expression microarrays (GSE101586, GSE101684, and GSE112214) were downloaded from Gene Expression Omnibus (GEO) database. Differentially expressed circRNAs (DECs) were obtained by the "limma" package of R programs and the overlapping operation was implemented of DECs. CircBase database and Cancer-Specific CircRNA database (CSCD) were used to find miRNAs binding to DECs. Target genes of the found miRNAs were identified utilizing Perl programs based on miRDB, miRTarBase, and TargetScan databases. Functional and enrichment analyses of selected target genes were performing using the "cluster profiler" package. Protein-protein interaction (PPI) network was constructed by the Search Tool for the STRING database and module analysis of selected hub genes was performed by Cytoscape 3.7.1. Survival analysis of hub genes were performed by Gene Expression Profiling Interactive Analysis (GEPIA). Respectively, 1 DEC, 249 DECs, and 101 DECs were identified in GSE101586, GSE101684, and GSE112214. A total of eight overlapped circRNAs, 43 miRNAs and 427 target genes were identified. Gene Ontology (GO) enrichment analysis showed these target genes were enriched in biological processes of regulation of histone methylation, Ras protein signal transduction and covalent chromatin modification etc. Pathway enrichment analysis showed these target genes are mainly involved in AMPK signaling pathway, signaling pathways regulating pluripotency of stem cells and insulin signaling pathway etc. A PPI network was constructed based on 427 target genes of the 43 miRNAs. Ten hub genes were found, of which the expression of MYLIP, GAN, and CDC27 were significantly related to NSCLC patient prognosis. Our study provide a deeper understanding the circRNAs-miRNAs-target genes by bioinformatics analysis, which may provide novel insights for unraveling pathogenesis of NSCLC. MYLIP, GAN, and CDC27 genes might serve as novel biomarker for precise treatment and prognosis of NSCLC in the future.
Keywords: bioinformatics; circRNA; expression; non-small cell lung cancer; pathway.
Copyright © 2020 Sun, Li, Xu, Zhang, Zuo, Xin, Zhang and Gong.
Figures





References
LinkOut - more resources
Full Text Sources
Research Materials

