Epitranscriptomic(N6-methyladenosine) Modification of Viral RNA and Virus-Host Interactions
- PMID: 33330128
- PMCID: PMC7732492
- DOI: 10.3389/fcimb.2020.584283
Epitranscriptomic(N6-methyladenosine) Modification of Viral RNA and Virus-Host Interactions
Abstract
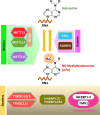
N6-methyladenosine (m6A) is the most prevalent and internal modification of eukaryotic mRNA. Multiple m6A methylation sites have been identified in the viral RNA genome and transcripts of DNA viruses in recent years. m6A modification is involved in all the phases of RNA metabolism, including RNA stability, splicing, nuclear exporting, RNA folding, translational modulation, and RNA degradation. Three protein groups, methyltransferases (m6A-writers), demethylases (m6A-erasers), and m6A-binding proteins (m6A-readers) regulate this dynamic reversible process. Here, we have reviewed the role of m6A modification dictating viral replication, morphogenesis, life cycle, and its contribution to disease progression. A better understanding of the m6A methylation process during viral pathogenesis is required to reveal novel approaches to combat the virus-associated diseases.
Keywords: m6A modification; m6A-binding protein; m6A-eraser; m6A-writer; viral epitranscriptomics.
Copyright © 2020 Imam, Kim and Siddiqui.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

