Histone Demethylase PHF8 Is Required for the Development of the Zebrafish Inner Ear and Posterior Lateral Line
- PMID: 33330448
- PMCID: PMC7719749
- DOI: 10.3389/fcell.2020.566504
Histone Demethylase PHF8 Is Required for the Development of the Zebrafish Inner Ear and Posterior Lateral Line
Abstract
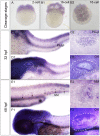
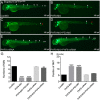
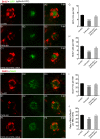
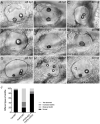
Histone demethylase PHF8 is crucial for multiple developmental processes, and hence, the awareness of its function in developing auditory organs needs to be increased. Using in situ hybridization (ISH) labeling, the mRNA expression of PHF8 in the zebrafish lateral line system and otic vesicle was monitored. The knockdown of PHF8 by morpholino significantly disrupted the development of the posterior lateral line system, which impacted cell migration and decreased the number of lateral line neuromasts. The knockdown of PHF8 also resulted in severe malformation of the semicircular canal and otoliths in terms of size, quantity, and position during the inner ear development. The loss of function of PHF8 also induced a defective differentiation in sensory hair cells in both lateral line neuromasts and the inner ear. ISH analysis of embryos that lacked PHF8 showed alterations in the expression of many target genes of several signaling pathways concerning cell migration and deposition, including the Wnt and FGF pathways. In summary, the current findings established PHF8 as a novel epigenetic element in developing auditory organs, rendering it a potential candidate for hearing loss therapy.
Keywords: PHF8; inner ear; organogenesis; posterior lateral line; zebrafish.
Copyright © 2020 He, Zheng, Luo, Hong, Su and Cai.
Figures




References
-
- David N. B., Sapede D., Saint-Etienne L., Thisse C., Thisse B., Dambly-Chaudiere C., et al. (2002). Molecular basis of cell migration in the fish lateral line: role of the chemokine receptor CXCR4 and of its ligand, SDF1. Proc. Natl. Acad. Sci. U.S.A. 99 16297–16302. 10.1073/pnas.252339399 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

