ANCA: A Web Server for Amino Acid Networks Construction and Analysis
- PMID: 33330622
- PMCID: PMC7711068
- DOI: 10.3389/fmolb.2020.582702
ANCA: A Web Server for Amino Acid Networks Construction and Analysis
Abstract
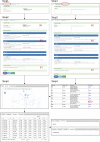
Amino acid network (AAN) models empower us to gain insights into protein structures and functions by describing a protein 3D structure as a graph, where nodes represent residues and edges as amino acid interactions. Here, we present the ANCA, an interactive Web server for Amino Acids Network Construction and Analysis based on a single structure or a set of structures from the Protein Data Bank. The main purpose of ANCA is to provide a portal for three types of an environment-dependent residue contact energy (ERCE)-based network model, including amino acid contact energy network (AACEN), node-weighted amino acid contact energy network (NACEN), and edge-weighted amino acid contact energy network (EACEN). For comparison, the C-alpha distance-based network model is also included, which can be extended to protein-DNA/RNA complexes. Then, the analyses of different types of AANs were performed and compared from node, edge, and network levels. The network and corresponding structure can be visualized directly in the browser. The ANCA enables researchers to investigate diverse concerns in the framework of AAN, such as the interpretation of allosteric regulation and functional residues. The ANCA portal, together with an extensive help, is available at http://sysbio.suda.edu.cn/anca/.
Keywords: ANCA portal; Amino acids network; allosteric regulation; functional residues; network analysis; protein structure.
Copyright © 2020 Yan, Yu, Chen, Zhou and Shen.
Figures
References
-
- Allaire J. J., Gandrud C., Russell K., Yetman C. J. (2017). “networkD3: D3 JavaScript Network Graphs From R”. R Package Version 0.4.
LinkOut - more resources
Full Text Sources