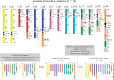
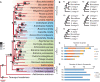
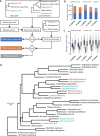
Linked by Ancestral Bonds: Multiple Whole-Genome Duplications and Reticulate Evolution in a Brassicaceae Tribe
- PMID: 33331908
- PMCID: PMC8097306
- DOI: 10.1093/molbev/msaa327
Linked by Ancestral Bonds: Multiple Whole-Genome Duplications and Reticulate Evolution in a Brassicaceae Tribe
Abstract
Pervasive hybridization and whole-genome duplications (WGDs) influenced genome evolution in several eukaryotic lineages. Although frequent and recurrent hybridizations may result in reticulate phylogenies, the evolutionary events underlying these reticulations, including detailed structure of the ancestral diploid and polyploid genomes, were only rarely reconstructed. Here, we elucidate the complex genomic history of a monophyletic clade from the mustard family (Brassicaceae), showing contentious relationships to the early-diverging clades of this model plant family. Genome evolution in the crucifer tribe Biscutelleae (∼60 species, 5 genera) was dominated by pervasive hybridizations and subsequent genome duplications. Diversification of an ancestral diploid genome into several divergent but crossable genomes was followed by hybridizations between these genomes. Whereas a single genus (Megadenia) remained diploid, the four remaining genera originated by allopolyploidy (Biscutella, Lunaria, Ricotia) or autopolyploidy (Heldreichia). The contentious relationships among the Biscutelleae genera, and between the tribe and other early diverged crucifer lineages, are best explained by close genomic relatedness among the recurrently hybridizing ancestral genomes. By using complementary cytogenomics and phylogenomics approaches, we demonstrate that the origin of a monophyletic plant clade can be more complex than a parsimonious assumption of a single WGD spurring postpolyploid cladogenesis. Instead, recurrent hybridization among the same and/or closely related parental genomes may phylogenetically interlink diploid and polyploid genomes despite the incidence of multiple independent WGDs. Our results provide new insights into evolution of early-diverging Brassicaceae lineages and elucidate challenges in resolving the contentious relationships within and between land plant lineages with pervasive hybridization and WGDs.
Keywords: chromosome rearrangements; diploidization; dysploidy; hybridization; phylogenetics; polyploidy; reticulate evolution; whole-genome duplication.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures




References
-
- Alexa A, Rahnenführer J, Lengauer T.. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22(13):1600–1607. - PubMed
-
- Al-Shehbaz IA. 2012. A generic and tribal synopsis of the Brassicaceae (Cruciferae). Taxon 61(5):931–954.
-
- Ané C, Larget B, Baum DA, Smith SD, Rokas A.. 2006. Bayesian estimation of concordance among gene trees. Mol Biol Evol. 24(2):412–426. - PubMed
-
- Barker MS, Dlugosch KM, Dinh L, Challa RS, Kane NC, King MG, Rieseberg LH.. 2010. EvoPipes.net: bioinformatic tools for ecological and evolutionary genomics. Evol Bioinform Online. 6:143–149. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

