SARS-CoV-2-specific T cells are rapidly expanded for therapeutic use and target conserved regions of the membrane protein
- PMID: 33331927
- PMCID: PMC7746091
- DOI: 10.1182/blood.2020008488
SARS-CoV-2-specific T cells are rapidly expanded for therapeutic use and target conserved regions of the membrane protein
Abstract
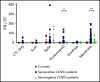
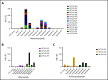
T-cell responses to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have been described in recovered patients, and may be important for immunity following infection and vaccination as well as for the development of an adoptive immunotherapy for the treatment of immunocompromised individuals. In this report, we demonstrate that SARS-CoV-2-specific T cells can be expanded from convalescent donors and recognize immunodominant viral epitopes in conserved regions of membrane, spike, and nucleocapsid. Following in vitro expansion using a good manufacturing practice-compliant methodology (designed to allow the rapid translation of this novel SARS-CoV-2 T-cell therapy to the clinic), membrane, spike, and nucleocapsid peptides elicited interferon-γ production, in 27 (59%), 12 (26%), and 10 (22%) convalescent donors (respectively), as well as in 2 of 15 unexposed controls. We identified multiple polyfunctional CD4-restricted T-cell epitopes within a highly conserved region of membrane protein, which induced polyfunctional T-cell responses, which may be critical for the development of effective vaccine and T-cell therapies. Hence, our study shows that SARS-CoV-2 directed T-cell immunotherapy targeting structural proteins, most importantly membrane protein, should be feasible for the prevention or early treatment of SARS-CoV-2 infection in immunocompromised patients with blood disorders or after bone marrow transplantation to achieve antiviral control while mitigating uncontrolled inflammation.
Conflict of interest statement
Conflict-of-interest disclosure: P.J.H. and C.R.C. are cofounders and are on the board of directors or scientific advisory board of Mana Therapeutics. P.J.H. is on a scientific advisory board for Cellevolve. C.R.C. is a consultant for Catamaran Bio. C.M.B. is on the advisory board for Cellectis and is cofounder and on the scientific advisory boards for Catamaran Bio and Mana Therapeutics with stock and/or ownership, is on the board of directors for Caballeta Bio with stock options and has stock in Neximmune and Torque Therapeutics. M.D.K. is on a scientific advisory panel for Gilead Sciences. K.M.H., M.D.K., C.A.L., P.J.H., C.R.C., A.A.A., and C.M.B. have filed a patent application based on the findings in this paper. The remaining authors declare no competing financial interests.
Figures





Comment in
-
Expanding the toolbox to combat a pandemic.Blood. 2020 Dec 17;136(25):2847-2848. doi: 10.1182/blood.2020009408. Blood. 2020. PMID: 33331932 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

