Recombination events are concentrated in the spike protein region of Betacoronaviruses
- PMID: 33332358
- PMCID: PMC7775116
- DOI: 10.1371/journal.pgen.1009272
Recombination events are concentrated in the spike protein region of Betacoronaviruses
Abstract
The Betacoronaviruses comprise multiple subgenera whose members have been implicated in human disease. As with SARS, MERS and now SARS-CoV-2, the origin and emergence of new variants are often attributed to events of recombination that alter host tropism or disease severity. In most cases, recombination has been detected by searches for excessively similar genomic regions in divergent strains; however, such analyses are complicated by the high mutation rates of RNA viruses, which can produce sequence similarities in distant strains by convergent mutations. By applying a genome-wide approach that examines the source of individual polymorphisms and that can be tested against null models in which recombination is absent and homoplasies can arise only by convergent mutations, we examine the extent and limits of recombination in Betacoronaviruses. We find that recombination accounts for nearly 40% of the polymorphisms circulating in populations and that gene exchange occurs almost exclusively among strains belonging to the same subgenus. Although experimental studies have shown that recombinational exchanges occur at random along the coronaviral genome, in nature, they are vastly overrepresented in regions controlling viral interaction with host cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


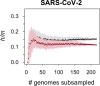
References
-
- Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, Xiang Z, Mu Z, Chen X, Chen J, Hu K, Jin Q, Wang J, Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020; 11(1):1620 10.1038/s41467-020-15562-9 - DOI - PMC - PubMed
-
- Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020; 579(7798):265–269. 10.1038/s41586-020-2008-3 - DOI - PMC - PubMed
-
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020; 579(7798):270–273. 10.1038/s41586-020-2012-7 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

