miRNA expression profiling and zeatin dynamic changes in a new model system of in vivo indirect regeneration of tomato
- PMID: 33332392
- PMCID: PMC7745965
- DOI: 10.1371/journal.pone.0237690
miRNA expression profiling and zeatin dynamic changes in a new model system of in vivo indirect regeneration of tomato
Abstract
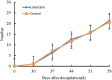
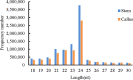
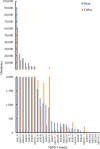
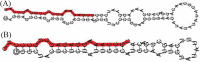
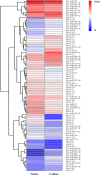
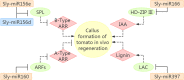
Callus formation and adventitious shoot differentiation could be observed on the cut surface of completely decapitated tomato plants. We propose that this process can be used as a model system to investigate the mechanisms that regulate indirect regeneration of higher plants without the addition of exogenous hormones. This study analyzed the patterns of trans-zeatin and miRNA expression during in vivo regeneration of tomato. Analysis of trans-zeatin revealed that the hormone cytokinin played an important role in in vivo regeneration of tomato. Among 183 miRNAs and 1168 predicted target genes sequences identified, 93 miRNAs and 505 potential targets were selected based on differential expression levels for further characterization. Expression patterns of six miRNAs, including sly-miR166, sly-miR167, sly-miR396, sly-miR397, novel 156, and novel 128, were further validated by qRT-PCR. We speculate that sly-miR156, sly-miR160, sly-miR166, and sly-miR397 play major roles in callus formation of tomato during in vivo regeneration by regulating cytokinin, IAA, and laccase levels. Overall, our microRNA sequence and target analyses of callus formation during in vivo regeneration of tomato provide novel insights into the regulation of regeneration in higher plants.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
MicroRNA Expression Profiles in Response to Phytophthora infestans and Oidium neolycopersici and Functional Identification of sly-miR397 in Tomato.Phytopathology. 2023 Mar;113(3):497-507. doi: 10.1094/PHYTO-04-22-0117-R. Epub 2023 Mar 24. Phytopathology. 2023. PMID: 36346372
-
Characterization and expression profiling of selected microRNAs in tomato (Solanum lycopersicon) 'Jiangshu14'.Mol Biol Rep. 2013 May;40(5):3503-21. doi: 10.1007/s11033-012-2425-y. Epub 2013 Feb 14. Mol Biol Rep. 2013. PMID: 23408149
-
Identification of Stress Associated microRNAs in Solanum lycopersicum by High-Throughput Sequencing.Genes (Basel). 2019 Jun 21;10(6):475. doi: 10.3390/genes10060475. Genes (Basel). 2019. PMID: 31234458 Free PMC article.
-
MicroRNAs in tomato plants.Sci China Life Sci. 2011 Jul;54(7):599-605. doi: 10.1007/s11427-011-4188-4. Epub 2011 Jul 12. Sci China Life Sci. 2011. PMID: 21748583 Review.
-
Harnessing spatial transcriptomics for advancing plant regeneration research.Trends Plant Sci. 2024 Jul;29(7):718-720. doi: 10.1016/j.tplants.2024.02.004. Epub 2024 Feb 28. Trends Plant Sci. 2024. PMID: 38418271 Review.
Cited by
-
Regulatory non-coding RNAs: Emerging roles during plant cell reprogramming and in vitro regeneration.Front Plant Sci. 2022 Nov 10;13:1049631. doi: 10.3389/fpls.2022.1049631. eCollection 2022. Front Plant Sci. 2022. PMID: 36438127 Free PMC article. Review.
-
Comprehensive regulatory networks for tomato organ development based on the genome and RNAome of MicroTom tomato.Hortic Res. 2023 Jul 19;10(9):uhad147. doi: 10.1093/hr/uhad147. eCollection 2023 Sep. Hortic Res. 2023. PMID: 37691964 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

