A TNXB splice donor site variant as a cause of hypermobility type Ehlers-Danlos syndrome in patients with congenital adrenal hyperplasia
- PMID: 33332743
- PMCID: PMC8077117
- DOI: 10.1002/mgg3.1556
A TNXB splice donor site variant as a cause of hypermobility type Ehlers-Danlos syndrome in patients with congenital adrenal hyperplasia
Abstract
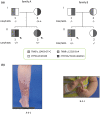
Background: Congenital adrenal hyperplasia (CAH) due to 21-hydroxylase deficiency is an autosomal recessive disease of steroidogenesis that affects 1 in 15,000. Approximately, 10% of the CAH population also suffer from CAH-X, a connective tissue dysplasia consistent with hypermobility type Ehlers-Danlos syndrome (EDS). Most patients with CAH-X carry a contiguous gene deletion involving CYP21A2 encoding 21-hydroxylase and TNXB encoding tenascin-X (TNX), but some are of unknown etiology.
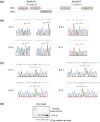
Methods: We conducted clinical evaluation and medical history review of EDS-related manifestations in subjects from two unrelated CAH families who carry a heterozygous TNXB c.12463+2T>C variant that alters the splice donor site of intron 42. A next generation sequencing (NGS) based EDS panel composed of 45 genes was performed for index patients from each family. TNX expression in patient skin biopsy tissues and dermal fibroblasts was assessed by qRT-PCR and Sanger sequencing.
Results: All three evaluated CAH patients carrying the TNXB splice site variant had moderate EDS manifestations. An NGS panel excluded involvement of other known EDS-related variants. RNA assay on skin biopsies and dermal fibroblasts did not detect splicing errors in TNX mRNA; however, the removal of intron 42 was less efficient in the allele harboring the splice site variant as evidenced by the existence of a premature TNX RNA form, leading to an allele specific decrease in TNX mRNA.
Conclusions: Carrying a TNXB c.12463+2T>C variant at the intron 42 splice donor site causes an allele specific decrease in TNX expression, which can be associated with moderate EDS in CAH patients.
Keywords: TNXB; CAH; EDS; Ehlers Danlos syndrome; congenital adrenal hyperplasia.
© 2021 The Authors. Molecular Genetics & Genomic Medicine published by Wiley Periodicals LLC. This article has been contributed to by US Government employees and their work is in the public domain in the USA.
Conflict of interest statement
D.P.M. received unrelated research funds from Diurnal Limited through the National Institutes of Health Cooperative Research and Development Agreement. The authors declare there is no conflict of interest in this work.
Figures

Similar articles
-
Measurement of serum tenascin-X in patients with congenital adrenal hyperplasia at risk for Ehlers-Danlos contiguous gene deletion syndrome CAH-X.BMC Res Notes. 2019 Oct 30;12(1):711. doi: 10.1186/s13104-019-4753-7. BMC Res Notes. 2019. PMID: 31666125 Free PMC article.
-
Tenascin-X haploinsufficiency associated with Ehlers-Danlos syndrome in patients with congenital adrenal hyperplasia.J Clin Endocrinol Metab. 2013 Feb;98(2):E379-87. doi: 10.1210/jc.2012-3148. Epub 2013 Jan 2. J Clin Endocrinol Metab. 2013. PMID: 23284009 Free PMC article.
-
Tenascin-X, Congenital Adrenal Hyperplasia, and the CAH-X Syndrome.Horm Res Paediatr. 2018;89(5):352-361. doi: 10.1159/000481911. Epub 2018 May 7. Horm Res Paediatr. 2018. PMID: 29734195 Free PMC article. Review.
-
Broadening the Spectrum of Ehlers Danlos Syndrome in Patients With Congenital Adrenal Hyperplasia.J Clin Endocrinol Metab. 2015 Aug;100(8):E1143-52. doi: 10.1210/jc.2015-2232. Epub 2015 Jun 15. J Clin Endocrinol Metab. 2015. PMID: 26075496 Free PMC article.
-
Congenital Adrenal Hyperplasia and Ehlers-Danlos Syndrome.Front Endocrinol (Lausanne). 2022 Feb 25;13:803226. doi: 10.3389/fendo.2022.803226. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35282436 Free PMC article. Review.
Cited by
-
DNA Methylation in Babies Born to Nonsmoking Mothers Exposed to Secondhand Smoke during Pregnancy: An Epigenome-Wide Association Study.Environ Health Perspect. 2021 May;129(5):57010. doi: 10.1289/EHP8099. Epub 2021 May 19. Environ Health Perspect. 2021. PMID: 34009014 Free PMC article.
-
Molecular basis and genetic testing strategies for diagnosing 21-hydroxylase deficiency, including CAH-X syndrome.Ann Pediatr Endocrinol Metab. 2023 Jun;28(2):77-86. doi: 10.6065/apem.2346108.054. Epub 2023 Jun 30. Ann Pediatr Endocrinol Metab. 2023. PMID: 37401054 Free PMC article.
-
Whole Exome Sequencing of 23 Multigeneration Idiopathic Scoliosis Families Reveals Enrichments in Cytoskeletal Variants, Suggests Highly Polygenic Disease.Genes (Basel). 2021 Jun 16;12(6):922. doi: 10.3390/genes12060922. Genes (Basel). 2021. PMID: 34208743 Free PMC article.
-
Molecular characterization of the new clinical entity associated with congenital adrenal hyperplasia: the CAH-X syndrome in the Spanish population.Adv Lab Med. 2023 Aug 25;4(3):258-267. doi: 10.1515/almed-2023-0071. eCollection 2023 Sep. Adv Lab Med. 2023. PMID: 38075167 Free PMC article.
-
Genes and Pseudogenes: Complexity of the RCCX Locus and Disease.Front Endocrinol (Lausanne). 2021 Jul 30;12:709758. doi: 10.3389/fendo.2021.709758. eCollection 2021. Front Endocrinol (Lausanne). 2021. PMID: 34394006 Free PMC article. Review.
References
-
- Aebi, M. , Hornig, H. , Padgett, R. A. , Reiser, J. , & Weissmann, C. (1986). Sequence requirements for splicing of higher eukaryotic nuclear pre‐mRNA. Cell, 47(4), 555–565. - PubMed
-
- Beighton, P. , De Paepe, A. , Steinmann, B. , Tsipouras, P. , & Wenstrup, R. J. (1998). Ehlers‐Danlos syndromes: Revised nosology, Villefranche, 1997. Ehlers‐Danlos National Foundation (USA) and Ehlers‐Danlos Support Group (UK). American Journal of Medical Genetics, 77(1), 31–37. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

