Targeted Protein Degradation Phenotypic Studies Using HaloTag CRISPR/Cas9 Endogenous Tagging Coupled with HaloPROTAC3
- PMID: 33332748
- PMCID: PMC7818660
- DOI: 10.1002/cpph.81
Targeted Protein Degradation Phenotypic Studies Using HaloTag CRISPR/Cas9 Endogenous Tagging Coupled with HaloPROTAC3
Abstract
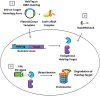
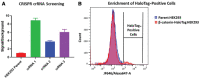
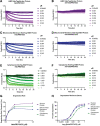
To assess the role of a protein, protein loss phenotypic studies can be used, most commonly through mutagenesis RNAi or CRISPR knockout. Such studies have been critical for the understanding of protein function and the identification of putative therapeutic targets for numerous human disease states. However, these methodological approaches present challenges because they are not easily reversible, and if an essential gene is targeted, an associated loss of cell viability can potentially hinder further studies. Here we present a reversible and conditional live-cell knockout strategy that is applicable to numerous proteins. This modular protein-tagging approach regulates target loss at the protein, rather than the genomic, level through the use of HaloPROTAC3, which specifically degrades HaloTag fusion proteins via recruitment of the VHL E3 ligase component. To enable HaloTag-mediated degradation of endogenous proteins, we provide protocols for HaloTag genomic insertion at the protein N or C terminus via CRISPR/Cas9 and use of HaloTag fluorescent ligands to enrich edited cells via Fluorescence-Activated Cell Sorting (FACS). Using these approaches, endogenous HaloTag fusion proteins present in various subcellular locations can be degraded by HaloPROTAC3. As detecting the degradation of endogenous targets is challenging, the 11-amino-acid peptide tag HiBiT is added to the HaloTag fusion to allows the sensitive luminescence detection of HaloTag fusion levels without the use of antibodies. Lastly, we demonstrate, through comparison of HaloPROTAC3 degradation with that of another fusion tag PROTAC, dTAG-13, that HaloPROTAC3 has a faster degradation rate and similar extent of degradation. © 2020 The Authors. Basic Protocol 1: CRISPR/Cas9 insertion of HaloTag or HiBiT-HaloTag Basic Protocol 2: HaloPROTAC3 degradation of endogenous HaloTag fusions.
Keywords: CRISPR; HaloPROTAC3; HaloTag; HiBiT; PROTAC; VHL; phenotype; targeted protein degradation.
© 2020 The Authors.
Figures


References
Literature Cited
-
- BasuRay, S. , Wang, Y. , Smagris, E. , Cohen, J. C. , & Hobbs, H. H. (2019). Accumulation of PNPLA3 on lipid droplets is the basis of associated hepatic steatosis. Proceedings of the National Academy of Sciences of the United States of America, 116(19), 9521–9526. doi: 10.1073/pnas.1901974116. - DOI - PMC - PubMed
-
- Bensimon, A. , Pizzagalli, M. D. , Kartnig, F. , Dvorak, V. , Essletzbichler, P. , Winter, G. E. , & Superti‐Furga, G. (2020). Targeted degradation of SLC transporters reveals amenability of multi‐pass transmembrane proteins to ligand‐induced proteolysis. Cell Chemical Biology, 27(6), 728–739.e729. doi: 10.1016/j.chembiol.2020.04.003. - DOI - PMC - PubMed
-
- Buckley, D. L. , Raina, K. , Darricarrere, N. , Hines, J. , Gustafson, J. L. , Smith, I. E. , … Crews, C. M. (2015). HaloPROTACS: Use of small molecule PROTACs to induce degradation of HaloTag fusion proteins. ACS Chemical Biology, 10(8), 1831–1837. doi: 10.1021/acschembio.5b00442. - DOI - PMC - PubMed
-
- Buckley, D. L. , Van Molle, I. , Gareiss, P. C. , Tae, H. S. , Michel, J. , Noblin, D. J. , … Crews, C. M. (2012). Targeting the von Hippel‐Lindau E3 ubiquitin ligase using small molecules to disrupt the VHL/HIF‐1alpha interaction. Journal of the American Chemical Society, 134(10), 4465–4468. doi: 10.1021/ja209924v. - DOI - PMC - PubMed
-
- Burslem, G. M. , Smith, B. E. , Lai, A. C. , Jaime‐Figueroa, S. , McQuaid, D. C. , Bondeson, D. P. , … Crews, C. M. (2018). The advantages of targeted protein degradation over inhibition: An RTK case study. Cell Chemical Biology, 25(1), 67–77.e63. doi: 10.1016/j.chembiol.2017.09.009. - DOI - PMC - PubMed
Internet Resources
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

