Genetic Screens Identify Host Factors for SARS-CoV-2 and Common Cold Coronaviruses
- PMID: 33333024
- PMCID: PMC7723770
- DOI: 10.1016/j.cell.2020.12.004
Genetic Screens Identify Host Factors for SARS-CoV-2 and Common Cold Coronaviruses
Abstract
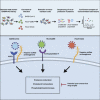
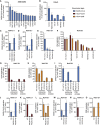
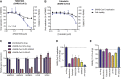
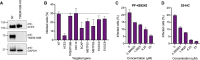
The Coronaviridae are a family of viruses that cause disease in humans ranging from mild respiratory infection to potentially lethal acute respiratory distress syndrome. Finding host factors common to multiple coronaviruses could facilitate the development of therapies to combat current and future coronavirus pandemics. Here, we conducted genome-wide CRISPR screens in cells infected by SARS-CoV-2 as well as two seasonally circulating common cold coronaviruses, OC43 and 229E. This approach correctly identified the distinct viral entry factors ACE2 (for SARS-CoV-2), aminopeptidase N (for 229E), and glycosaminoglycans (for OC43). Additionally, we identified phosphatidylinositol phosphate biosynthesis and cholesterol homeostasis as critical host pathways supporting infection by all three coronaviruses. By contrast, the lysosomal protein TMEM106B appeared unique to SARS-CoV-2 infection. Pharmacological inhibition of phosphatidylinositol kinases and cholesterol homeostasis reduced replication of all three coronaviruses. These findings offer important insights for the understanding of the coronavirus life cycle and the development of host-directed therapies.
Keywords: 229E; COVID-19; CRISPR; OC43; SARS-CoV-2; coronavirus; genetic screen; host factors; host-targeted antivirals; virus-host interactions.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests J.C.-S., J.O., and K.H. are employees of Synthego Corporation. All authors declare no other competing interests.
Figures







Update of
-
Functional genomic screens identify human host factors for SARS-CoV-2 and common cold coronaviruses.bioRxiv [Preprint]. 2020 Sep 24:2020.09.24.312298. doi: 10.1101/2020.09.24.312298. bioRxiv. 2020. Update in: Cell. 2021 Jan 7;184(1):106-119.e14. doi: 10.1016/j.cell.2020.12.004. PMID: 32995787 Free PMC article. Updated. Preprint.
Comment in
-
A Crisp(r) New Perspective on SARS-CoV-2 Biology.Cell. 2021 Jan 7;184(1):15-17. doi: 10.1016/j.cell.2020.12.003. Epub 2020 Dec 17. Cell. 2021. PMID: 33338422 Free PMC article.
References
-
- Baggen J., Persoons L., Jansen S., Vanstreels E., Jacquemyn M., Jochmans D., Neyts J., Dallmeier K., Maes P., Daelemans D. Identification of TMEM106B as proviral host factor for SARS-CoV-2. BioRxiv. 2020 doi: 10.1101/2020.09.28.316281. - DOI
-
- Balderhaar H.J., Ungermann C. CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion. J. Cell Sci. 2013;126:1307–1316. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 AI060537/AI/NIAID NIH HHS/United States
- R01 AI120694/AI/NIAID NIH HHS/United States
- T32 GM007618/GM/NIGMS NIH HHS/United States
- U19 AI135990/AI/NIAID NIH HHS/United States
- R01 AI122747/AI/NIAID NIH HHS/United States
- F32 CA239333/CA/NCI NIH HHS/United States
- R01 AI141970/AI/NIAID NIH HHS/United States
- R01 AI130123/AI/NIAID NIH HHS/United States
- DP1 DA038043/DA/NIDA NIH HHS/United States
- U19 AI135972/AI/NIAID NIH HHS/United States
- R01 AI143292/AI/NIAID NIH HHS/United States
- P01 AI063302/AI/NIAID NIH HHS/United States
- R01 AI140186/AI/NIAID NIH HHS/United States
- P50 AI150476/AI/NIAID NIH HHS/United States
- T32 GM136534/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

