Synergistic and Antagonistic Drug Combinations against SARS-CoV-2
- PMID: 33333292
- PMCID: PMC7834738
- DOI: 10.1016/j.ymthe.2020.12.016
Synergistic and Antagonistic Drug Combinations against SARS-CoV-2
Abstract
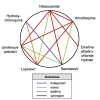
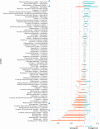
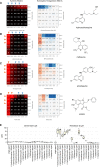
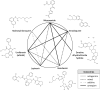
Antiviral drug development for coronavirus disease 2019 (COVID-19) is occurring at an unprecedented pace, yet there are still limited therapeutic options for treating this disease. We hypothesized that combining drugs with independent mechanisms of action could result in synergy against SARS-CoV-2, thus generating better antiviral efficacy. Using in silico approaches, we prioritized 73 combinations of 32 drugs with potential activity against SARS-CoV-2 and then tested them in vitro. Sixteen synergistic and eight antagonistic combinations were identified; among 16 synergistic cases, combinations of the US Food and Drug Administration (FDA)-approved drug nitazoxanide with remdesivir, amodiaquine, or umifenovir were most notable, all exhibiting significant synergy against SARS-CoV-2 in a cell model. However, the combination of remdesivir and lysosomotropic drugs, such as hydroxychloroquine, demonstrated strong antagonism. Overall, these results highlight the utility of drug repurposing and preclinical testing of drug combinations for discovering potential therapies to treat COVID-19.
Keywords: COVID-19; CPE assay; SARS-CoV-2; combination therapy; drug combinations; drug repurposing; drug synergy; in silico design; knowledge mining; nitazoxanide remdesivir combo.
Copyright © 2020 The American Society of Gene and Cell Therapy. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures


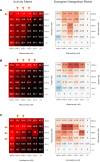
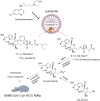
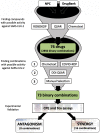
Update of
-
Discovery of Synergistic and Antagonistic Drug Combinations against SARS-CoV-2 In Vitro.bioRxiv [Preprint]. 2020 Jun 30:2020.06.29.178889. doi: 10.1101/2020.06.29.178889. bioRxiv. 2020. Update in: Mol Ther. 2021 Feb 3;29(2):873-885. doi: 10.1016/j.ymthe.2020.12.016. PMID: 32637956 Free PMC article. Updated. Preprint.
References
-
- Chou T.C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006;58:621–681. - PubMed
-
- Sun X., Vilar S., Tatonetti N.P. High-Throughput Methods for Combinatorial Drug Discovery. Sci. Transl. Med. 2013;5:205rv1. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

