Comprehensive Functional Characterization and Clinical Interpretation of 20 Splice-Site Variants of the RAD51C Gene
- PMID: 33333735
- PMCID: PMC7765170
- DOI: 10.3390/cancers12123771
Comprehensive Functional Characterization and Clinical Interpretation of 20 Splice-Site Variants of the RAD51C Gene
Abstract
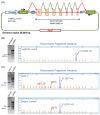
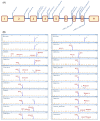
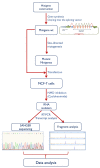
Hereditary breast and/or ovarian cancer is a highly heterogeneous disease with more than 10 known disease-associated genes. In the framework of the BRIDGES project (Breast Cancer Risk after Diagnostic Gene Sequencing), the RAD51C gene has been sequenced in 60,466 breast cancer patients and 53,461 controls. We aimed at functionally characterizing all the identified genetic variants that are predicted to disrupt the splicing process. Forty RAD51C variants of the intron-exon boundaries were bioinformatically analyzed, 20 of which were selected for splicing functional assays. To test them, a splicing reporter minigene with exons 2 to 8 was designed and constructed. This minigene generated a full-length transcript of the expected size (1062 nucleotides), sequence, and structure (Vector exon V1- RAD51C exons_2-8- Vector exon V2). The 20 candidate variants were genetically engineered into the wild type minigene and functionally assayed in MCF-7 cells. Nineteen variants (95%) impaired splicing, while 18 of them produced severe splicing anomalies. At least 35 transcripts were generated by the mutant minigenes: 16 protein-truncating, 6 in-frame, and 13 minor uncharacterized isoforms. According to ACMG/AMP-based standards, 15 variants could be classified as pathogenic or likely pathogenic variants: c.404G > A, c.405-6T > A, c.571 + 4A > G, c.571 + 5G > A, c.572-1G > T, c.705G > T, c.706-2A > C, c.706-2A > G, c.837 + 2T > C, c.905-3C > G, c.905-2A > C, c.905-2_905-1del, c.965 + 5G > A, c.1026 + 5_1026 + 7del, and c.1026 + 5G > T.
Keywords: RAD51C; VUS; aberrant splicing; breast cancer; clinical interpretation; functional assay; genetic variants; minigene; ovarian cancer; splicing; susceptibility genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Schubert S., Luttikhuizen J.L., Auber B., Schmidt G., Hofmann W., Penkert J., Davenport C.F., Hille-Betz U., Wendeburg L., Bublitz J., et al. The identification of pathogenic variants in BRCA1/2 negative, high risk, hereditary breast and/or ovarian cancer patients: High frequency of FANCM pathogenic variants. Int. J. Cancer. 2019;144:2683–2694. doi: 10.1002/ijc.31992. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

