Non-Coding RNAs as Prognostic Biomarkers: A miRNA Signature Specific for Aggressive Early-Stage Lung Adenocarcinomas
- PMID: 33333738
- PMCID: PMC7768474
- DOI: 10.3390/ncrna6040048
Non-Coding RNAs as Prognostic Biomarkers: A miRNA Signature Specific for Aggressive Early-Stage Lung Adenocarcinomas
Abstract
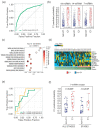
Lung cancer burden can be reduced by adopting primary and secondary prevention strategies such as anti-smoking campaigns and low-dose CT screening for high risk subjects (aged >50 and smokers >30 packs/year). Recent CT screening trials demonstrated a stage-shift towards earlier stage lung cancer and reduction of mortality (~20%). However, a sizable fraction of patients (30-50%) with early stage disease still experience relapse and an adverse prognosis. Thus, the identification of effective prognostic biomarkers in stage I lung cancer is nowadays paramount. Here, we applied a multi-tiered approach relying on coupled RNA-seq and miRNA-seq data analysis of a large cohort of lung cancer patients (TCGA-LUAD, n = 510), which enabled us to identify prognostic miRNA signatures in stage I lung adenocarcinoma. Such signatures showed high accuracy (AUC ranging between 0.79 and 0.85) in scoring aggressive disease. Importantly, using a network-based approach we rewired miRNA-mRNA regulatory networks, identifying a minimal signature of 7 miRNAs, which was validated in a cohort of FFPE lung adenocarcinoma samples (CSS, n = 44) and controls a variety of genes overlapping with cancer relevant pathways. Our results further demonstrate the reliability of miRNA-based biomarkers for lung cancer prognostication and make a step forward to the application of miRNA biomarkers in the clinical routine.
Keywords: biomarkers; gene expression; lung cancer; microRNA; prognosis.
Conflict of interest statement
Authors declare no conflict of interest.
Figures


References
-
- Travis W.D., Colby T.V., Corrin B., Shimosato Y., Brambilla E. Histological Typing of Lung and Pleural Tumours. 3rd ed. Springer; Berlin/Heidelberg, Germany: 1999. WHO. World Health Organization. International Histological Classification of Tumours.
Grants and funding
LinkOut - more resources
Full Text Sources

