Identification, Phylogeny, and Comparative Expression of the Lipoxygenase Gene Family of the Aquatic Duckweed, Spirodela polyrhiza, during Growth and in Response to Methyl Jasmonate and Salt
- PMID: 33333747
- PMCID: PMC7765210
- DOI: 10.3390/ijms21249527
Identification, Phylogeny, and Comparative Expression of the Lipoxygenase Gene Family of the Aquatic Duckweed, Spirodela polyrhiza, during Growth and in Response to Methyl Jasmonate and Salt
Abstract
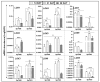
Lipoxygenases (LOXs) (EC 1.13.11.12) catalyze the oxygenation of fatty acids and produce oxylipins, including the plant hormone jasmonic acid (JA) and its methyl ester, methyl jasmonate (MeJA). Little information is available about the LOX gene family in aquatic plants. We identified a novel LOX gene family comprising nine LOX genes in the aquatic plant Spirodela polyrhiza (greater duckweed). The reduced anatomy of S. polyrhiza did not lead to a reduction in LOX family genes. The 13-LOX subfamily, with seven genes, predominates, while the 9-LOX subfamily is reduced to two genes, an opposite trend from known LOX families of other plant species. As the 13-LOX subfamily is associated with the synthesis of JA/MeJA, its predominance in the Spirodela genome raises the possibility of a higher requirement for the hormone in the aquatic plant. JA-/MeJA-based feedback regulation during culture aging as well as the induction of LOX gene family members within 6 h of salt exposure are demonstrated.
Keywords: MeJA; Spirodela polyrhiza; duckweed; lipoxygenases (LOXs); oxylipin; phylogenetics; salt.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Hillman W.S. The Lemnaceae, or duckweeds. Bot. Rev. 1961;27:221–287. doi: 10.1007/BF02860083. - DOI
-
- Bog M., Appenroth K.-J., Sree K.S. Duckweed (Lemnaceae): Its Molecular Taxonomy. Front. Sustain. Food Syst. 2019;3:117. doi: 10.3389/fsufs.2019.00117. - DOI
-
- Landolt E. The Family of Lemnaceae—A Monographic Study (Vol. 1) Volume 71. Geobotanische Institut ETH, Stiftung Rübel; Zurich, Switzerland: 1986. p. 566.
-
- Wang W., Kerstetter R., Michael T. Evolution of genome size in duckweeds (Lemnaceae) J. Bot. 2011;2:1. doi: 10.1155/2011/570319. - DOI
-
- Mattoo A.K., Pick U., Hoffman-Falk H., Edelman M. The rapidly metabolized 32,000-dalton polypeptide of the chloroplast is the “proteinaceous shield” regulating photosystem II electron transport and mediating diuron herbicide sensitivity. Proc. Natl. Acad. Sci. USA. 1981;78:1572–1576. doi: 10.1073/pnas.78.3.1572. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

